Full Text
After nearly 30 years of experience with offshore wind energy (OWE), the industry is moving past the initial learning phase and into large-scale development. One of the strongest motivations for OWE is that projects are large enough to replace fossil-fueled electricity production that we know is a primary contributor to the ongoing climate crisis. Environmental concerns are therefore also at the core of OWE development, and environmental research and impact monitoring have been central parts of the industry since its inception. A large volume of science-based knowledge is available about the environmental impacts of OWE on marine ecosystems, from effects on the seafloor sediment infauna beneath the turbines to those on marine mammals that roam the developed seas.
In the next couple of years, we will see a large increase in OWE development, with single projects such as Dogger Bank in the North Sea coming online as well as larger projects that produce several gigawatts of energy from thousands of square kilometers devoted to OWE production. Because they can be deployed in deep water, floating wind turbines play a significant role in these long-term developmental plans, but their environmental impacts are not well understood, including the possible impacts on pelagic ecosystems. With large projects covering ever more area, including deep waters, we may see cumulative effects on upper ocean stratification as projected by oceanographic models of lee effects (Broström, 2008). Turbulence introduced by the foundation structures and anchoring lines may add to the forces that result in increased mixing of upper water layers with potential downstream consequences on productivity (Floeter et al., 2017).
Another knowledge gap concerns the general impact on pelagic fish from floating OWE. Trawl surveys are typically used to monitor pelagic fish; however, trawling is difficult in areas with large structures such as turbine foundations and their anchor lines. While demersal species of fish are relatively well researched in OWE installations, often using passive gear such as bottom set gill nets or fyke nets, the impact of the structures on behavior and distribution of pelagic species is less known (Methratta et al., 2020).
To address these knowledge gaps and assess a new method for impact monitoring at large offshore wind installations, we developed and tested a strategy for sampling environmental DNA in the pelagic ecosystem surrounding a floating OWE installation (Figure 1). Environmental DNA (or eDNA) is DNA shed from animals and plants to the soil, air, sediment, or water environment where it can be sampled by filtration with pore sizes in the range of one micrometer or smaller. The captured eDNA can be assessed in various ways. Here, we wanted to assess species composition of pelagic metazoan communities, with an emphasis on fish species. To accomplish this, we used metabarcoding with the relatively slowly evolving gene 18S (V1–V2), which amplifies a range of eukaryote taxa, and the fish-specific MiFish assay based on a fragment of the faster evolving, species-specific 12S mitochondrial gene. We also wanted to estimate the abundances of two of the commercially most important pelagic species in the area, Atlantic herring (Clupea harengus) and Atlantic mackerel (Scomber scombrus). For these quantitative analyses we used herring- and mackerel-specific primers in assays optimized for droplet digital PCR (ddPCR), a technology that permits estimates of the number of target gene copies in a sample.
|
|
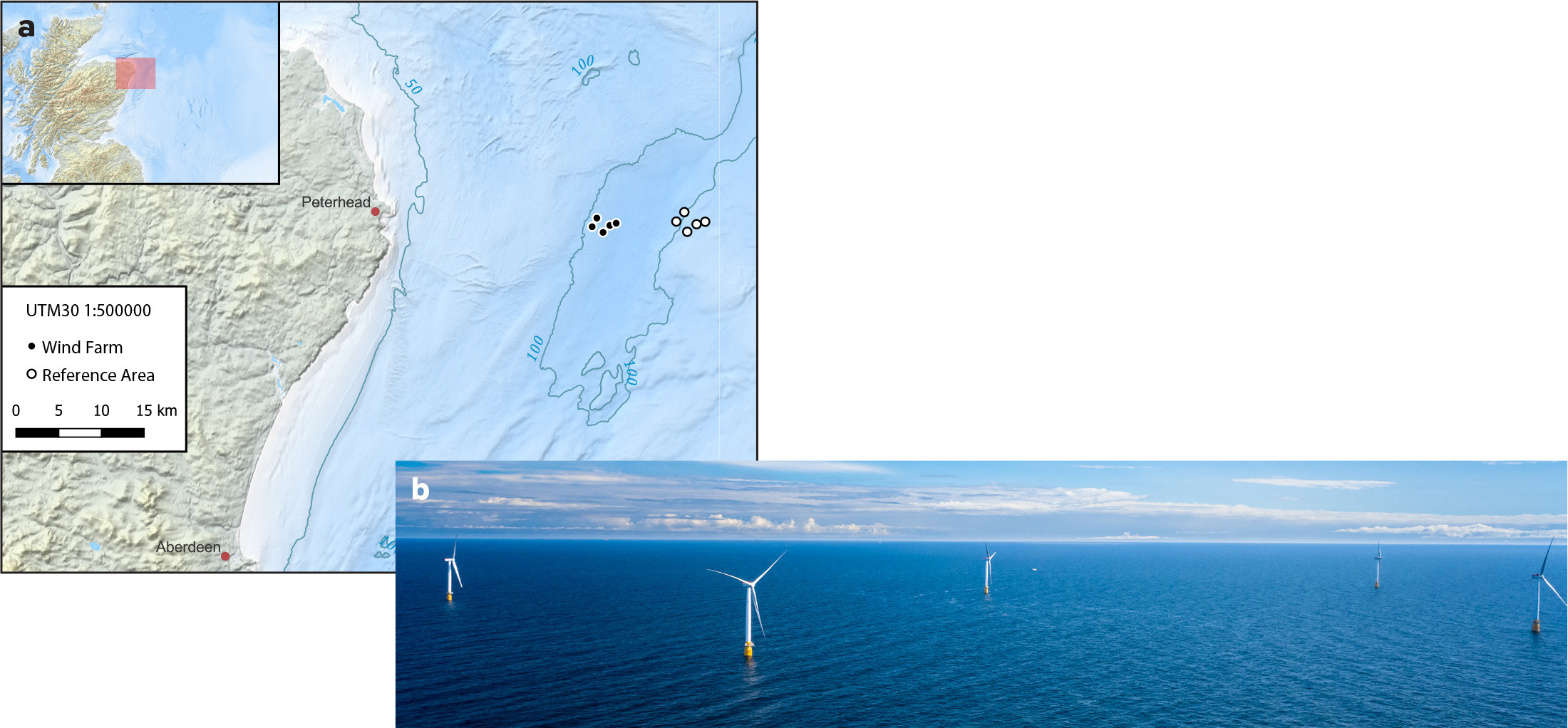
The test was carried out in August 2021 at Equinor’s pilot installation Hywind Scotland, situated 25 km east of Peterhead in northeastern Scotland and operational since 2017 (Figure 1a). The wind farm consists of five floating 6 MW turbines installed at 95–120 m depth, spaced 800–1,600 m apart, and attached to the seafloor with three-point mooring systems (Figure 1b). We used a similar sized reference area with the same depth range located approximately 10 km east of the wind farm. Five stations in each of these areas were sampled at 10 m and 50 m depths. Each sample consisted of 3 × 1 liter of water. The samples were immediately filtered and preserved on board and kept dark and cool to reduce eDNA decay prior to sample analysis.
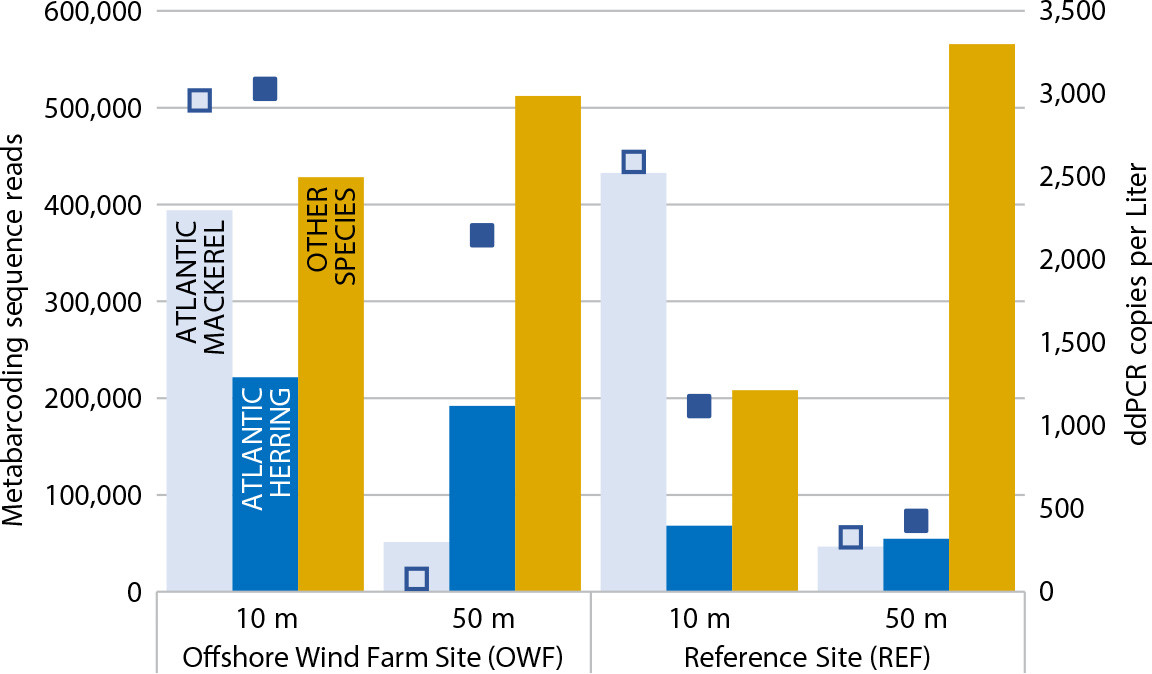
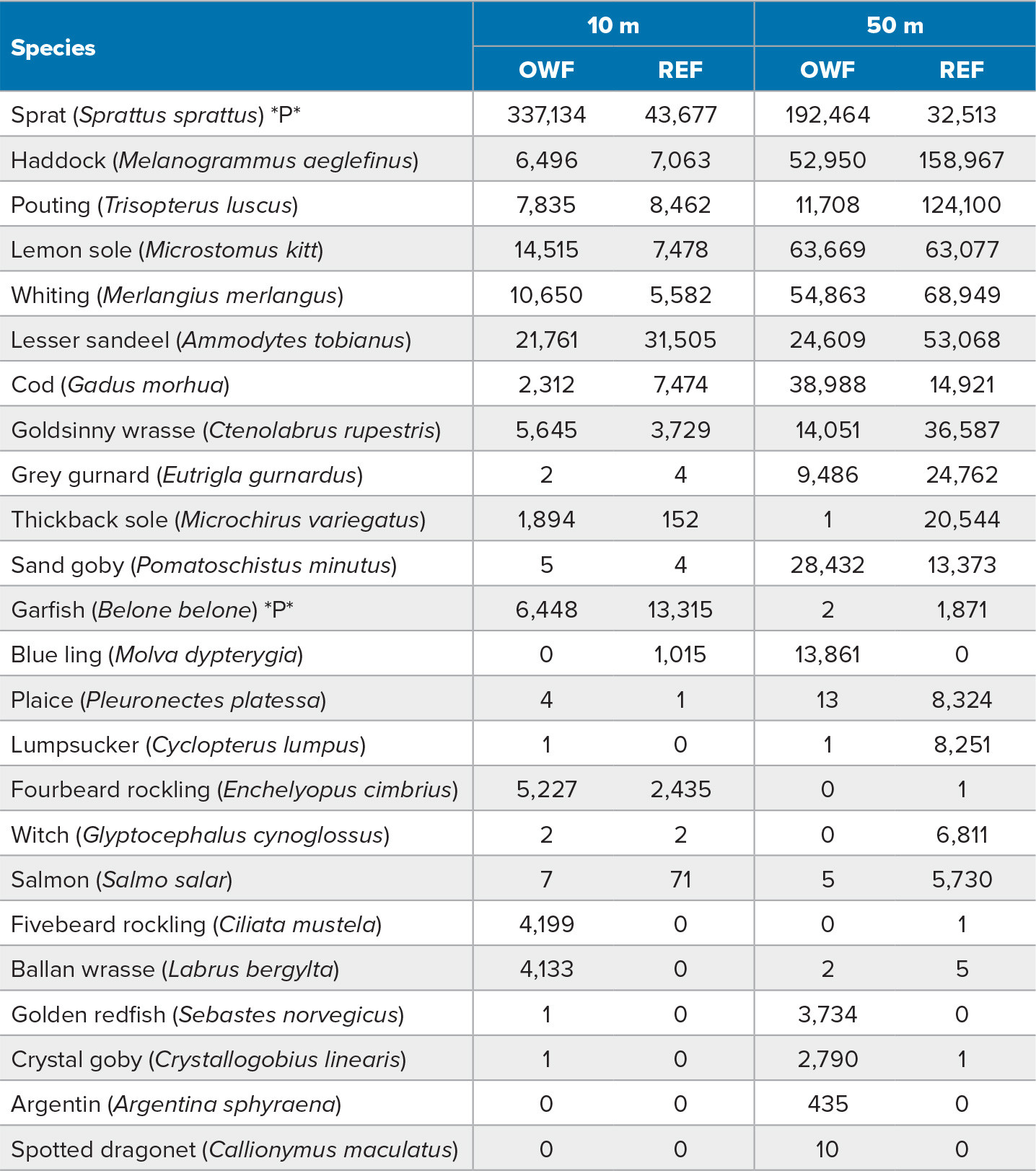
The most striking result from the fish community composition analysis, using MiFish metabarcoding data, was the strong signal from demersal species (Figure 2). While the highest abundances were indicated for the three pelagic species herring, sprat, and mackerel, the data also included at least 22 bottom-dwelling species (Table S1). The signal was highest at 50 m depth, but some demersal species were also detected at 10 m. There was no significant difference between the installation and the reference area, but the two depths were consistently different from each other in the 20 sampled stations. While not a target species in this analysis, a faint eDNA signal from harbor porpoise was detected in the MiFish data from one of the wind-farm stations. Previous reports suggest that porpoises actively seek out offshore wind installations for foraging (Fernandez-Betelu et al., 2022). This shows that optimizing eDNA markers for marine mammals could facilitate inclusion of such species in future eDNA surveys.
|
|
TABLE S1. Species detected in Hywind Scotland eDNA MiFish metabarcoding excluding Atlantic herring and Atlantic mackerel presented in Figure 2. The number of reads for each species is given at each of the two depths (10 m and 50 m) sampled for the offshore wind farm site (OWF) and the reference site (REF). Pelagic species are marked with a *P*. > High res table |
Sampling was completed on one day only, so any attracting or repelling effects from the turbine foundations and mooring lines may have been masked by variations caused by dynamically moving schools of fish. More conclusive results would require sampling over time, including collecting diurnal and seasonal data. No signals from the target species were detected in relatively large fractions of ddPCR analyses, indicating that it would be advantageous to use larger volumes of filtered water than the 1 liter chosen for this study to limit time spent in the field. Given these caveats, species-specific abundance estimates (ddPCR data) indicated a higher abundance of herring in the wind farm relative to the reference site (Figure 2). This was true for both the 10 m and 50 m sampled depths. The mackerel data indicated a higher abundance at 10 m than 50 m in both areas, with no difference between the areas. Both results were supported by the number of reads from the metabarcoding data (Figure 2).
In our eukaryotic metabarcoding analysis, protists comprised the majority of sequence reads. As with the MiFish data, cluster analyses of the 18S data indicate no difference between the two areas, but there is a strong clustering of data based on depth. As this study represents a snapshot at a specific point in time, and plankton typically migrate vertically in the water column over a 24-hour period, a sampling campaign would ideally extend over at least one full migratory cycle.
Our multi-pronged eDNA approach offers a powerful tool for identifying spatial and temporal structuring of pelagic communities. This will facilitate monitoring of change in hard-to-reach areas such as OWE installations where traditional methods are difficult to use. It may also increase the possibilities for identifying ecosystem changes at regional scales and over a broad range of taxons in a way that traditional sampling cannot do.
Pairing metabarcoding for broad-scale community level assessment with ddPCR to obtain quantitative information regarding important target species is a powerful approach for characterizing the structures of pelagic communities.