INTRODUCTION
Ocean species, especially those with large populations, tend to occur across broad geographic ranges, and as a result they inhabit a variety of physical and chemical environments. For species that are highly mobile as adults, the combination of wide ranges and environmental variation results in populations that occupy different habitat types during their lives (Block et al., 2011). For species with more sedentary adults, different subpopulations can inhabit very different environments. This pattern is particularly clear for the many coastal marine taxa that produce mobile, dispersing larvae (Jackson, 1974), including the majority of commercially important coastal species (Bradbury et al., 2008).
Living across an environmental mosaic is particularly common in temperate coastal species that can often range from subtropical to subarctic latitudes (Roy et al., 1998). Such habitats often have strong temperature gradients, show latitudinal shifts in growing season start or growing season length, and are based on a variety of geologies, from rocky coasts to marshes to estuarine regions. Along the US west coast, there is also a strong shift in coastal upwelling, with upwelling hotspots dotting the waters from California to Washington State (Feely et al., 2008). Upwelling brings cold, low pH, low oxygenated seawater to shallow coastal zones, dramatically changing rates of growth, feeding, and metabolism plus a host of other basic features of physiology (M. Kelly and Hoffman, 2013; Somero et al., 2016; Low and Micheli, 2018). These upwelling centers, layered along the temperature and seasonal gradients from subtropical Baja California to subarctic Alaska, produce a wide range of habitats, many of which are likely to shift with the continuation of climate change: as temperatures warm, upwelling centers change, and oxygen levels drop (see other articles in this special issue, plus Deutsch et al., 2015).
As environments shift, populations face natural selection that can lead to increased adaptation to local conditions. This kind of evolution during climate change can help determine the dynamics of future populations and their degrees of range shift (e.g., Pinsky et al., 2013). As a result, the nature of local adaptation to the environment is an important component of predicting future marine population ecology (Sanford and Kelly, 2011), and can help to protect the high levels of adaptive genetic variation that are essential to future climate adaptation (Norberg et al., 2012). Highly dispersive marine larvae create substantial gene flow in many coastal species, moving larvae into environmental patches that are different from those of their parents. By contrast, larvae with lower movement, such as the crawl-away larvae of some gastropods or the short-term swimming larvae of ascidians, tend to settle near their parents and grow in similar environmental conditions (Sanford and Kelly, 2011). As a result, the potential to adapt to local environments is quite different for these two types of species. In fact, the key comparison is the dispersal range of a species per generation compared to the size of environmental patches. When environmental patches are large compared to dispersal capacity (Figure 1a), then offspring largely settle into similar environments and adaptation can proceed from generation to generation. Genes that might be selected by one environment versus another can differ across these environments (represented by ovals of different color in Figure 1). However, when environmental patch size is small compared to dispersal, then offspring settle from a larval pool into different environments. Natural selection can occur in these settings, but acts as a single-cohort filter of existing variation.
Genes that are adaptive in one environment might be selected against in another, resulting in genetic differentiation across habitat boundaries (Figure 1b). But this differentiation is set by the balance between gene flow and selection in each generation. In the next generation, larvae from different habitats may again mix, leading to a high degree of genetic polymorphism in the larval pool (Levene, 1953). As a result, study of the evolution and environmental physiology of marine species across an environmental mosaic is expected to uncover a plethora of genetic variation for adaptive traits, structured over scales that depend on the strength of selection, its geographic scale, and the scale of dispersal.
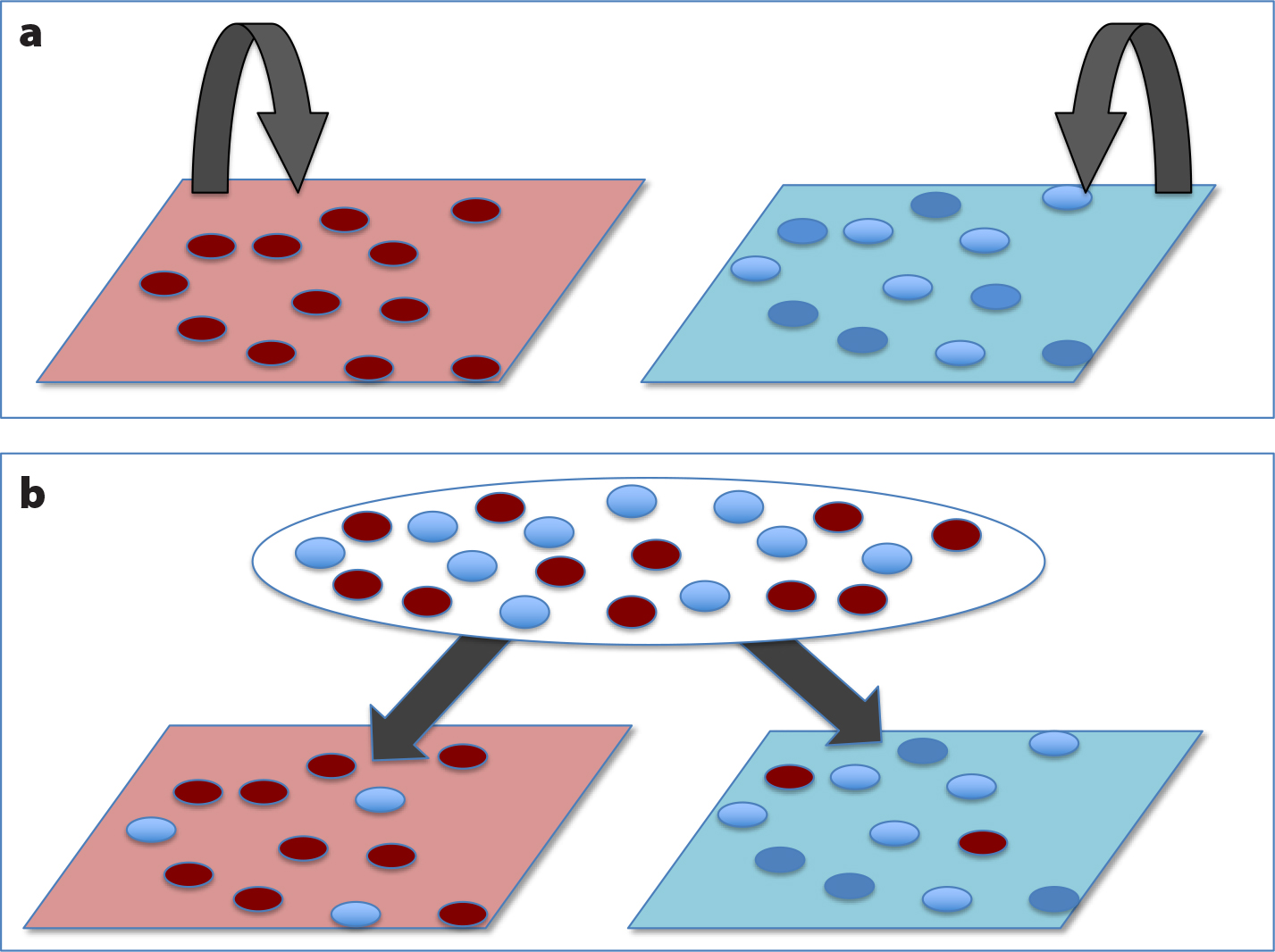
FIGURE 1. Local adaptation, dispersal, and environmental patch size. When different environmental patches (red vs. blue panels in (a)) are large compared to dispersal (arrows), natural selection among different genotypes (red vs. blue circles) leads to local adaptation over many generations. When patches are small compared to dispersal (b), then larvae from a larval pool settle in different patch types and selection can act only one generation at a time. This one-generation-at-a-time selection can lead to differences between patches, but also tends to retain genetic variation within and between patches. > High res figure
|
Along the US west coast, patterns of spatial variation in nearshore environmental conditions have been extensively studied, including temperature, pH, and oxygen, coupled with monitoring of abundance and population dynamics of a range of invertebrate and fish species (see other articles in this special issue). Because dispersal occurs in a mobile larval phase for many of these species, patterns of local adaptation may be slight or substantial, depending on how strongly selection acts and how variable the genes (and their phenotypic effects) are. This research demands a multi-level focus—from broad issues in marine conservation and ecology to deep understanding of the physiological, genetic, biochemical, and molecular factors that can ultimately determine if and how well an organism can function in different habitats.
PHYSIOLOGY INTEGRATES ENVIRONMENTAL IMPACTS TO CHANGE POPULATION DYNAMICS AND EVOLUTION
Changes in the physical and chemical makeup of seawater, whether it be shifts in temperature, oxygen concentration, or pH, ultimately impact marine organisms through effects on physiology. These and other environmental variables influence processes that are fundamental to maintaining homeostasis, meaning that changing ocean conditions exert strong selection pressure for marine populations to adjust physiology in order to survive and reproduce (Somero et al., 2016, 2017). The results generate changes in demography, population growth, species interactions, and species ranges that together influence broad-scale ecosystem functioning. As a result, the ongoing dynamics of physical change, physiological adjustment, and population response create strong linkages from physical science to ecology to evolution. But these links are not one way. Evolution can change how species react, and changes in species abundance can alter the sensitivity of ecosystem processes to environmental change. Climate change acts to alter the physical makeup of ocean environments, and so ripples through the hierarchy of biological responses.
Below, we offer five examples of studies that have used recent advances in genome technology to obtain insights into the patterns and mechanisms of local adaptation across the US west coast. The goals of these studies range from demonstrating some of the ways species adapt to their local conditions to showing that local adaptation can occur within single species despite substantial dispersal. Different studies (protein based, transcriptome based, gene frequency based) illustrate myriad ways for identifying the genomic and proteomic mechanisms by which local adaptation to climate change-relevant environmental conditions occurs. The geographic setting of the US west coast offers the possibility of studying a large array of species across strong gradients in temperature, upwelling, productivity, oxygen, settlement density, and more. A collaborative analysis of ocean chemistry, physics, environmental evolution, and ecology along the US west coast, such as the one developed by the Partnership for Interdisciplinary Study of the Coastal Oceans (PISCO), can set the stage for understanding how species and populations have adapted to current environments and how they may change in the future. Such studies range from examination of single gene adaptations of similar species to temperature to genomic investigations of the responses of populations to acidification, considerations of broader genomic views of multi-gene adaptation along the coast, and multi-species compilations of genetic structures and species assemblages. They chronicle a consistent pattern of local adaptation over small and large scales, and show a host of genetic resources that are available for responding to present and future climate.
MECHANISM OF ADAPTATION: STRUCTURE-FUNCTION RELATIONSHIPS IN THE ENZYME MALATE DEHYDROGENASE (MDH)
One of the foci of molecular level studies has been the adaptation of various species to different thermal conditions. Coastal habitats often differ widely in temperature. For example, at a given rocky intertidal site, animals living high up on rocks may experience temperatures near 40°C, whereas subtidal species living less than a meter away may seldom, if ever, encounter body temperatures higher than about 15°C. How, then, do the thermal sensitivities differ among species that occur at different heights and latitudes and help to explain the distribution patterns of similar species?
Proteins—which are responsible for much of organisms’ basic structures and physiological activities—are extremely sensitive to even small changes in temperature (Somero et al., 2017). As a result, targeted studies of protein function have been very useful in detecting species and even population differentiation. The best-studied protein for addressing issues of temperature adaptation is the enzyme cytosolic malate dehydrogenase (cMDH), which plays a critical role in generating a cell’s energy supply. Detailed studies have shown that the structural stabilities of cMDH proteins reflect adaptation to the different body temperatures found among species due to latitude and vertical position (Dong and Somero, 2009; Dong et al., 2018; Liao et al., 2019). As predicted, the higher the body temperature of a species, the more resistant the enzyme to loss of activity during heating (heat denaturation). This trend is shown for cMDH proteins from snails found at different heights in the rocky intertidal zone at Hopkins Marine Station (Figure 2): snails living higher in the intertidal heat up more at low tide and have more heat resistant cMDH. This strong positive correlation between body temperature and protein thermal stability has been observed across a wide range of taxa from different latitudes (Liao et al., 2019), and is characteristic of all classes of enzymes examined to date (Fields et al., 2015; Somero et al., 2017).
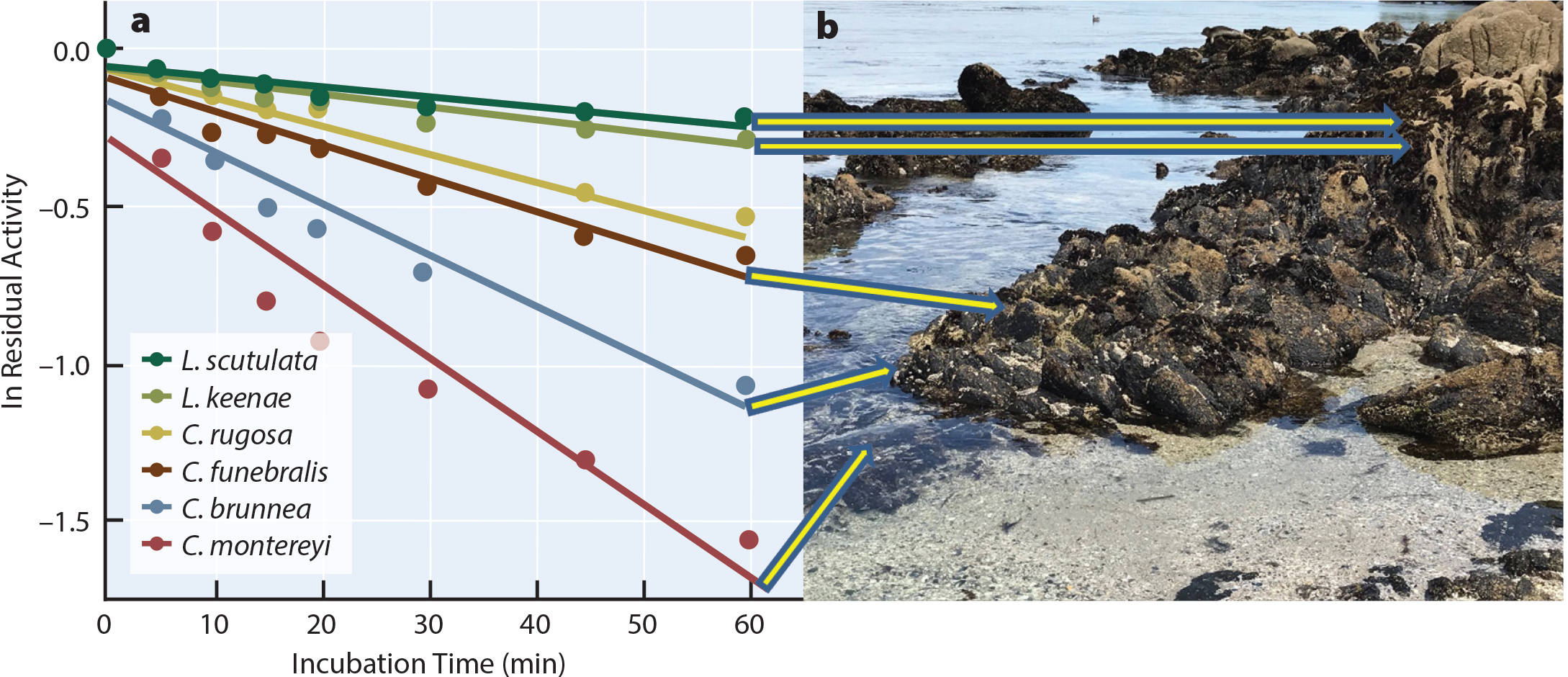
FIGURE 2. Variation in thermal stability among cytosolic-malate dehydrogenase (cMDH) enzymes from marine snails. (a) Loss of activity (indexed by ln of residual [remaining] activity) during heating at 42.5°C. cMDHs are from four species of turban snails (genus Chlorostoma [formerly Tegula]) from different vertical sites and two species of Littorina snails (note that C. rugosa is a low-intertidal species from a different location, the Gulf of California, and so does not have a line to its intertidal zone). (b) The rocky intertidal zone at Hopkins Marine Station. Figure modified after Somero et al. (2017). > High res figure
|
This adaptive fine-tuning of temperature sensitivities is not restricted to overall protein stability. cMDHs work by binding to substrate molecules and then converting them to metabolic products to keep the cell’s energy production going at needed rates. Substrate binding is intrinsically stronger in high temperature species, so that at their high body temperatures, enzymes of these species maintain binding abilities similar to those of orthologous enzymes (variants of a protein encoded by the same gene in all species) of low temperature species. This ensures that biochemical function can be sustained at normal body temperatures even as those temperatures vary (Somero et al., 2017; Liao et al., 2019).
These studies of cMDH orthologs have also suggested answers to several additional questions about enzyme-temperature relationships. First, how much change in the ~330 amino acid sequence of a cMDH protein is needed to alter thermal sensitivity of structure and function? Second, where in the protein structure are these changes concentrated? Third, how, mechanistically, do these changes in sequence bring about adaptive change in stability and function? The answer to the first question initially came from studies of two closely related congeners of the limpet genus Lottia, L. digitalis, a more northerly occurring species, and L. austrodigitalis, a more southerly occurring species (Dong and Somero, 2009). The temperature-adaptive differences between the cMDH orthologs were due to only a single amino acid substitution, one that led to enhanced internal bonding (stability) in the more warm-adapted ortholog. Further comparative studies of cMDHs from other species in different temperature settings confirmed this pattern: only minimal sequence differences are sufficient to achieve adaptation to temperature if the change occurs at the right spot in the protein (Liao et al., 2019).
In this case, the “right spot” turns out to be one of several sites in the sequence that influence the flexibility of the regions of the enzyme whose movements are critical for function. Enzymes are machines with moving parts, and the movements of the parts must occur in a precise manner for proper function, notably the binding of substrate molecules and the release of reaction products. In general, amino acid changes that occur during evolution at different temperatures modulate the stabilities of these moving parts, allowing them to retain the correct three-dimensional shapes needed for function, yet to maintain the flexibility to undergo essential movements associated with binding of substrates and the release of products (Dong et al., 2018; Liao et al., 2019).
The findings that, firstly, only one to a few changes in sequence are adequate to achieve adaptation to temperature, and secondly, that several sites in the sequence can be altered to allow adaptation have implications for the speed with which organisms’ responses to global change might occur. Thus, if each protein requires only minimal “fine-tuning” to keep up with warming, and if the locations of this “tuning” independently involve separate sites in the sequence, adaptive responses to rising temperatures may be relatively feasible—if enough genetic variation exists in populations. In the case of Lottia, transcriptome sequences of individuals co-occurring in Monterey Bay show high polymorphism: nearly 100,000 single nucleotide polymorphisms occur across 8,100 genes, including five polymorphisms in malate dehydrogenase genes (author Palumbi, unpublished data). Future studies of genetic variation, and of the requirements for adaptation by other types of proteins whose thermal sensitivities may differ from cMDH, may provide us with clearer estimates of how ongoing increases in temperature will affect latitudinal and vertical patterning of marine communities. In this broad biogeographical context, it merits noting that the northward spread up the west coast of North America of an invasive mussel native to the Mediterranean Sea, Mytilus galloprovincialis, may be due in part to the invasive’s more warm-adapted physiology relative to the native congener it is replacing, M. trossulus (Lockwood and Somero, 2011, 2012). A wide range of physiological, biochemical, and molecular differences, including differences in cMDH thermal sensitivities, distinguish these two species. A warming climate may shift the relative competitive abilities of these two congeners and lead to this type of biogeographic shift. Thus, molecular-level studies may provide insights not only into the abilities of native species to withstand climate change and remain in their current habitats, but also provide insight into habitats that these species may invade in the future.
ACIDIFICATION AS AN ADAPTIVE CHALLENGE
A range of genomic tools also makes it possible to study adaptation within species. This is particularly relevant to studies of environmental stress caused by climate change. Two of the least controversial aspects of climate science are that CO2 is increasing in the global atmosphere (Keeling, 2008) and that higher levels of CO2 are entering the ocean at the sea/air interface (Sabine et al., 2004). Higher CO2 in turn leads to higher levels of carbonic acid, and thus to higher levels of acidity (= lower pH). Because pH regulation is a very conserved feature in cellular physiology, and because the calcium carbonate skeletons of many marine species are often susceptible to low pH (Kroeker et al., 2010), future pH conditions are likely to be an important driver of ocean ecosystems (Feely et al., 2008).
Determining the range of pH conditions currently encountered by marine populations is the first step toward understanding the physiological effects of ocean acidification, and recently developed sensors provide fine-grain resolution of pH dynamics in nearshore marine communities along the US west coast (Martz et al., 2010; Hofmann et al., 2011; Chan et al., 2017). These pH measurements reveal a strong geographic pattern that is persistent across multiple years of observations (Figure 3a,b). Regional centers of acidification occur where ocean upwelling brings deep, cold, low pH water close to the surface (Feely et al., 2008). This spatial pattern of pH variation indicates that populations of broadly distributed species will encounter markedly different frequencies of low pH exposure and that the strength of selection for low pH tolerance will vary across this region.
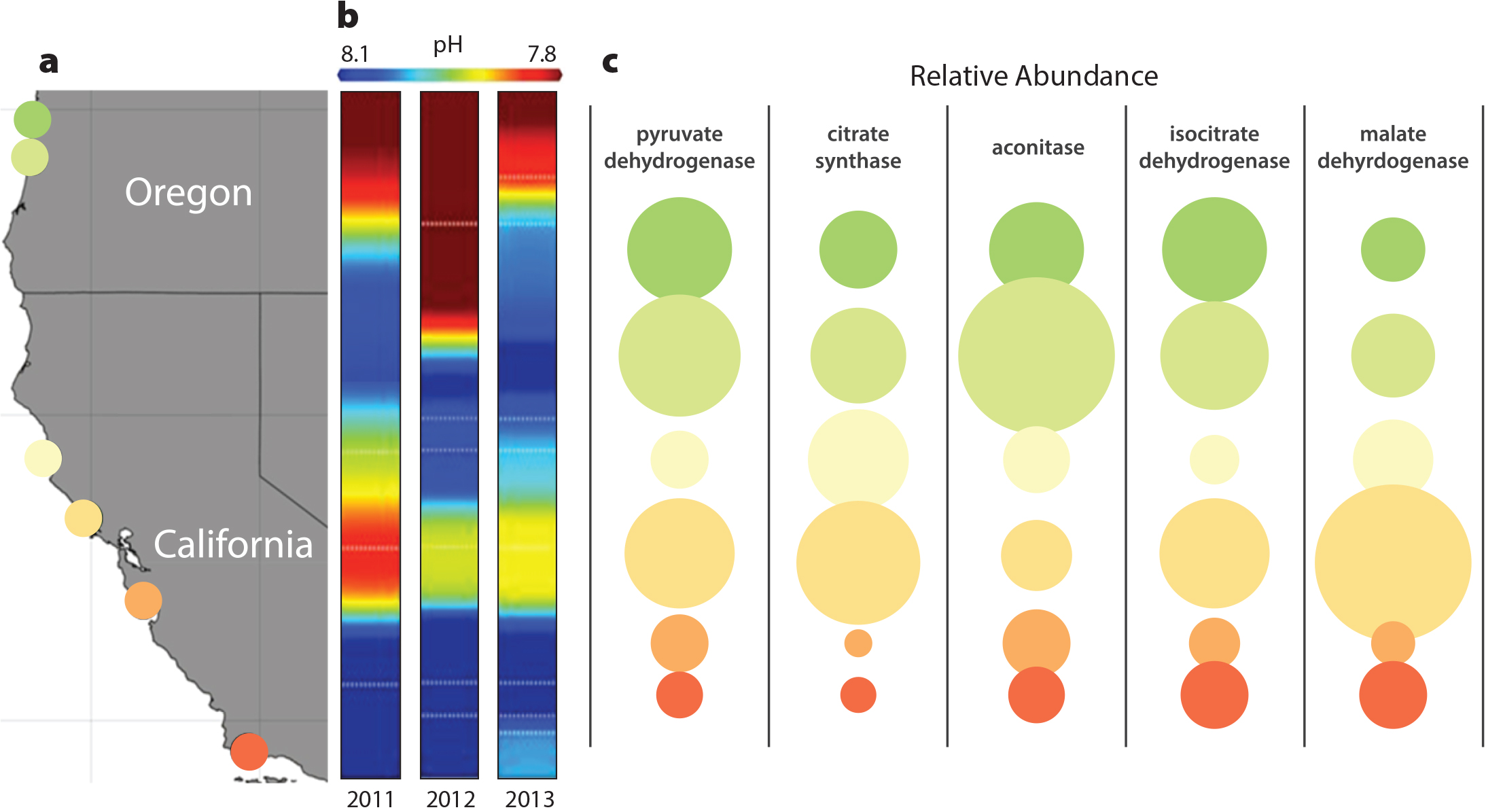
FIGURE 3. (a) Location of pH sensors and S. purpuratus sampling locations. Adapted from Evans et al. (2017) (b) Severity of low pH exposure across three years of pH sensor deployment. Reproduced from Chan et al. (2017) under Creative Commons license (http://creativecommons.org/licenses/by/4.0/) (c) Relative abundance of mRNAs encoding five proteins within the citric acid cycle following low pH exposure among S. purpuratus populations. Adapted from Evans et al. (2017). Graphic by M. Kovacs. > High res figure
|
Understanding how this pH mosaic has affected the physiology of ocean species is an important question (Evans et al., 2015), and the purple sea urchin (Strongylocentrotus purpuratus) provides a useful study system for this research. Purple sea urchin abundance has a strong influence on the overall structure of nearshore marine communities: high urchin densities create intense grazing pressure that can transform highly productive kelp forests into so-called “urchin barrens” that are far less biodiverse (Pearse, 2006). Purple urchins have highly dispersive larvae that can be transported widely across latitudes, implying that larvae will often drift to areas where the degree of acidification is considerably different than that encountered by their parents. Populations of larvae that have successfully settled and survived in low pH centers may have been selected for physiological mechanisms of tolerance to low pH that are less common in populations residing in higher pH areas where selection pressure is reversed or relaxed (M. Kelly et al., 2013; Pespeni et al., 2013a,c).
Experiments that compare gene expression response to seawater acidification among urchin populations from different pH regimes serve to identify physiological pathways acted on by natural selection to promote homeostasis in seawater with different pH. By measuring changes in the abundance of messenger RNA (mRNA), a precursor to the proteins that perform most of the physiological work inside cells, Evans et al. (2017) demonstrate that purple urchin larvae spawned from adults living in low pH centers respond differently to seawater acidification than larvae from parents residing in areas of comparatively high pH. Close inspection of mRNAs from approximately 20,000 S. purpuratus genes indicates that larvae from low pH populations increase the abundance of mRNAs involved in energy production, particularly within the citric acid cycle (a series of chemical reactions, in which cMDH plays a key role, involved in transforming carbohydrates, lipids, and proteins into cellular fuel and building blocks for structures; Figure 3c). These data suggest that the metabolic physiology of these urchins is fine-tuned to local pH conditions and that tolerance of low pH seawater is mediated in part by energy generation through the citric acid cycle (Evans et al., 2017).
Maintaining pH homeostasis is an energetically demanding process. Hydrogen ions responsible for reduced pH diffuse from seawater into the extracellular spaces of marine organisms and disrupt acid-base balance (Stumpp et al., 2012). High concentration of hydrogen ions near sites of calcification can prevent urchins from properly forming the hard spines they need for swimming, feeding, and defense (Byrne et al., 2013). Restoring calcification may involve increasing the activity of transport proteins that pump hydrogen ions away from calcification sites and other extracellular spaces, a process that requires considerable energy input (Pan et al., 2015). Pespeni et al. (2013b) showed that northern urchins from areas of persistent, strong upwelling and low pH in Oregon grew spines at a slower rate than those from southern California, even when sea urchins from these two locations were grown for a year in the same conditions. The complex adaptations that govern skeletal growth under variable conditions of temperature and pH are under study in a wide range of systems (e.g., the skate Leucoraja erinacea; Di Santo, 2019).
Increased tolerance of low pH seawater may represent a double-edged sword for purple sea urchins. On one hand, the capacity to tolerate pH conditions considered extreme relative to other marine environments indicates that these urchins may be well equipped for life in the future ocean. On the other hand, enhanced low pH tolerance may come with a trade-off: increased energy expenditure and a higher overall “cost of living.” In fact, the Levene (1953) model predicts that genetic polymorphisms that generate such trade-offs persist longer in the population because some alleles fare better in one kind of condition, and the other alleles are the opposite. How such trade-offs will affect the long-term population dynamics of purple sea urchins, or other marine species forced to adapt to a changing ocean through similar energy-intensive mechanisms, remains uncertain. However, these uncertainties provide new and exciting opportunities for integrated research.
THE POPULATION GENOMICS OF A PHYSIOLOGICALLY TRACTABLE MODEL SYSTEM
The studies discussed above focus on specific adaptive changes in a well-known species subject to experimental manipulation in the lab. They reveal patterns of physiological- and molecular-level change that suggest local adaptation is a powerful force in even high dispersal marine species. These data suggest we should search for other adaptive changes across marine populations as a way to gauge the importance of natural selection in crafting marine population dynamics over large spatial scales. Partitioning of genetic variants—alleles—in space can reveal population structure and identify potential targets of selection. Comparing genome-wide genetic variation partitioned between urchins from Oregon and San Diego, Pespeni et al. (2010) revealed a consistent signature of higher genetic differentiation than expected under neutrality in a specific set of genes related to immunity, regulation of protein production, reproduction, and development. This examination of genetic variation in 17,000 genes also found an excess of heterozygosity in immune-related genes, particularly in urchins from San Diego, potentially due to selection that enhances genetic diversity because of the greater incidence of disease and pathogen abundance in the southern California Bight region (Pespeni et al., 2012).
Comparison of putative adaptive and neutral loci among six populations from Bamfield, Canada, to Baja California, Mexico, showed strong correlations between population allele frequencies and local temperatures, while controlling for latitude, in putative adaptive loci and not in neutral loci (Pespeni and Palumbi, 2013). These results provided strong evidence that locally adapted alleles can be maintained via natural selection despite the homogenizing effects of gene flow (Pespeni and Palumbi, 2013), and they expanded the list of physiological systems likely to be acted on by natural selection in the context of environmental change.
As genome-level studies have become more common, strong selection acting on a few genes has become more and more visible. Such genetic changes at a few loci are often embedded in widespread genetic homogeneity across other loci. For example, red abalone living within strong upwelling zones subject to strong acidification showed differentiation from neighboring populations outside upwelling zones at a few loci involved in shell deposition and growth (De Wit and Palumbi, 2013). Yet, these populations showed no differentiation across thousands of other genes, consistent with previous microsatellite data. Homogeneity across most loci suggests consistent gene flow along the coast, a result typically used to inform fisheries management. By contrast, differentiation at a few loci suggests the action of strong local natural selection even in the presence of dispersal. These differentiated populations are still demographically connected to others along the coast, but face distinct environmental and ecological challenges. As a result, the use of population genomics as a tool to monitor connectivity now provides the opportunity to distinguish dispersal-driven gene flow and environmental variation strong enough to impact local population growth, such as in efforts to disentangle dispersal and selection in populations of Atlantic cod (Barth et al., 2017).
TESTING THE GENETIC “LOTTERY” HYPOTHESIS FOR LARVAL SELECTION
When natural selection acts within a set of settling marine larvae, it can generate changes in allele frequencies that result in one-generation evolutionary change. These genetic lotteries depend on input of adaptive genetic variation that provides higher fitness to some settlers than to others in that local environment. To determine whether populations of purple sea urchins exhibit genetic variation that would help them survive future ocean acidification conditions, and whether strong selection could generate genetic change in one generation, Pespeni et al. (2013a) developed a method for identifying alleles responsive to specific selective treatments using single generation selection experiments. They measured changes in allele frequency through developmental time for sea urchin larvae reared in ambient (400 ppm) and elevated (900 ppm) CO2 partial pressures (pCO2) for seven purple urchin populations from sites along the upwelling mosaic from Oregon to Santa Barbara, California. They found that larvae raised in high pCO2 conditions showed rapid allelic shifts through time in genes related to lipid metabolism and ion homeostasis, two functional classes of genes with important roles for responding to pH stress (Pespeni et al., 2013a). These results suggest selection favored alleles that improved performance under the high pCO2 treatment via the maintenance of physiological and morphological homeostasis during development.
In addition, alleles that responded to experimental acidification were also highly differentiated across local pH conditions in the wild, suggesting that both natural and experimental acidification targeted the same genes in selecting physiological phenotypes that perform better in low pH conditions (Pespeni et al., 2013c; Figure 4). Taken together, these results suggest that heterogeneity in pH conditions in the wild due to the natural process of upwelling promotes the maintenance of genetic variation that is adaptive in future ocean acidification conditions. However, rapid genetic response comes with a cost; mortality even within one generation results in a lower larval population size and a decrease in genetic diversity in the high pCO2 environment (Lloyd et al., 2016).
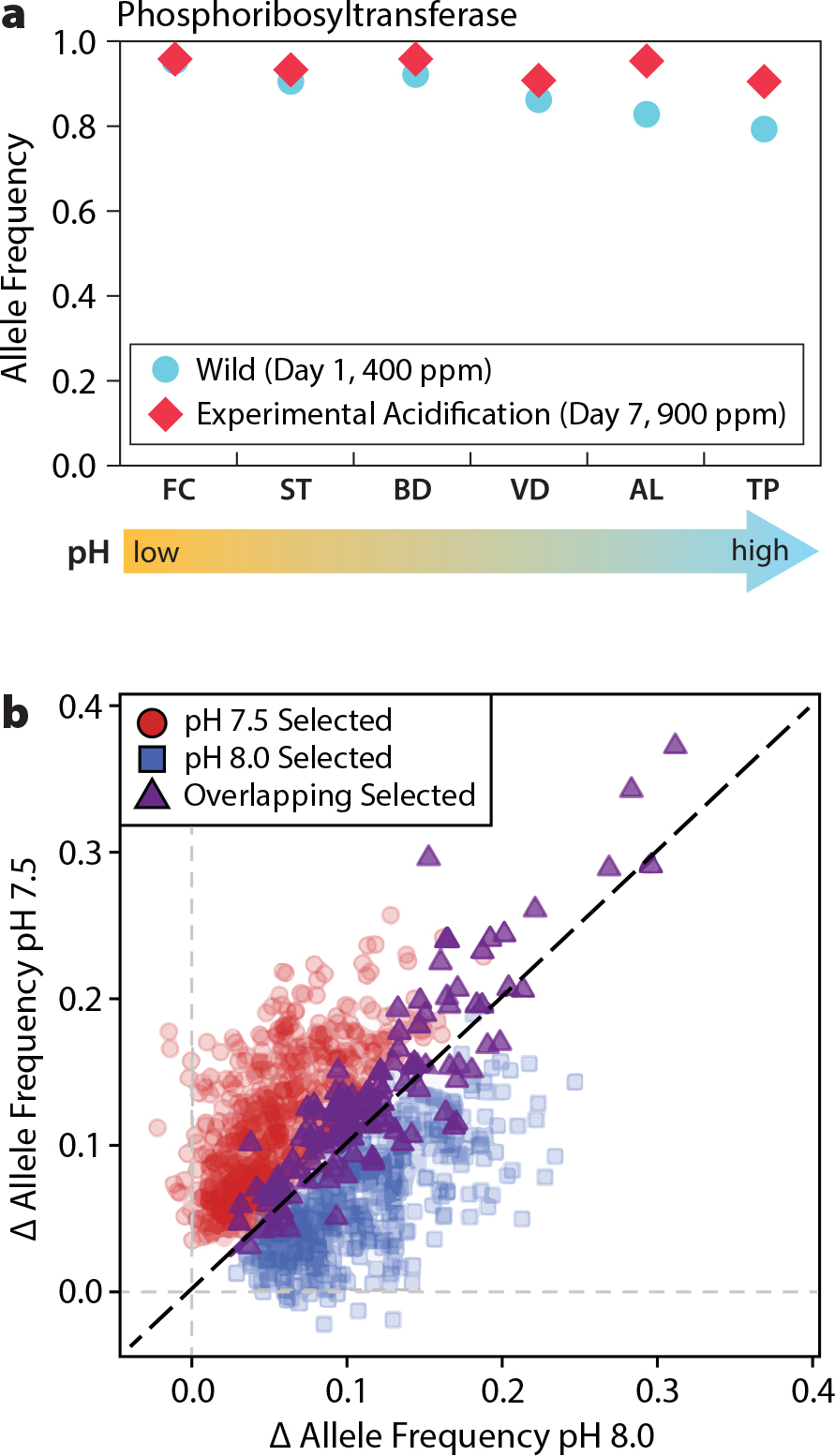
FIGURE 4. pH-adaptive genetic variation in purple sea urchin populations. (a) Allele frequencies of putative low-pH adaptive alleles correlate with local pH conditions in the wild (blue circles) and show greater changes in allele frequency toward the putative low-pH adaptive allele in response to experimental acidification (red diamonds). Populations sampled from Oregon to southern California are ordered in this figure by the amount of time spent at pH <7.8. FC = Fogarty Creek, ST = Strawberry Hill, BD = Bodega, VD = Van Damme State Park, AL = Alegria, Santa Barbara, and TP = Terrace Point, Santa Cruz. Modified from Pespeni et al. (2013c) (b) Correlated changes in allele frequency in moderate and extreme low-pH responsive alleles. From Brennan et al. (2019). > High res figure
|
The combination of natural upwelling “hotspots” and anthropogenic ocean acidification results in future ocean pH conditions that are predicted to be more variable with more frequent extreme low pH events (Feely et al., 2008; Pacella et al., 2018). The experimental acidification in the studies above utilized a low pH treatment within the bounds of what is frequently experienced by purple sea urchins in nature, pH 7.8 (900 ppm pCO2; Hofmann et al., 2011; Evans et al., 2013, 2015). Rearing larvae in moderate (pH 8.0) and extreme (pH 7.5) low pH conditions, Brennan et al. (2019) found purple urchin larvae to have adaptive standing genetic variation for survival in extreme low pH conditions that show correlated responses to moderate low pH conditions (Figure 4; Brennan et al., 2019). Interestingly, two classes of genetic variation emerge as useful for survival in low pH conditions: (1) variants that respond to both pH conditions and are maintained at high frequency in natural populations, and (2) relatively rare genetic variants that respond to either pH condition (Brennan et al., 2019). These results emphasize (1) the importance of environmental variation that potentially leads to balancing selection in the maintenance of common adaptive variants (Levene, 1953), and (2) the importance of large population sizes for the maintenance of rare genetic variation that is adaptive in global change conditions.
THE GENETIC MAP OF A COASTLINE
A picture of genetic variation along the west coast of North America has developed from studies described above that include work to collect very specific knowledge of protein thermal adaptation between species, detailed physiological experiments based on specific acidification conditions, and a broad, genome-wide look at genetic adaptation of a single species. The next step is a broad-based examination of the variation and genetic structure of a wide range of fish and invertebrate species along this coast, with the goal of understanding the scope for adaptation within species as well as the distribution of this genetic variation at the ecosystem scale.
A comparison of 50 invertebrate species using mitochondrial DNA (mtDNA) sequences showed abundant genetic diversity in all species, and listed 28 species without coastal genetic differentiation based on this assay (R. Kelly and Palumbi, 2010). The occurrence of strong structure was greatest between Alaska and Oregon, moderate between Oregon and Monterey (across Cape Mendocino), and low between Monterey and Santa Barbara (across Point Conception). Thus, the important biogeographic barrier (at the species level) at Point Conception is less of a genetic barrier for the species in this assay (Figure 5). Data for fish also show high levels of genetic variation and significant geographic patterns: genetic structure is strong in nine rockfish species that are concentrated along the northern coast from Oregon to Monterey and along the southern coast from Monterey to Santa Barbara (Figure 6). The fish data suggest stronger structure in more species, but this may be an artifact of differences in the data sets. The fish data include mtDNA and numerous nuclear genes, and so are likely to reveal more fine-grained patterns than the invertebrate surveys based on mtDNA alone.
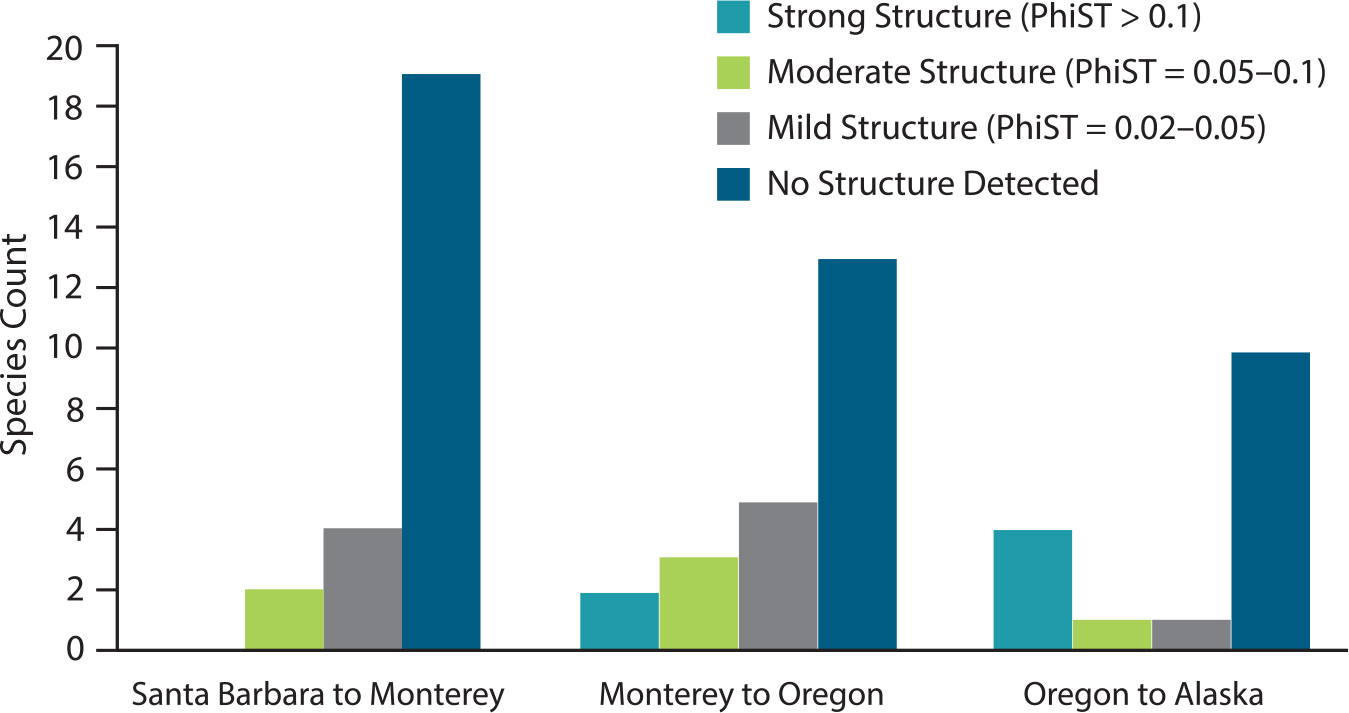
FIGURE 5. The geographic distribution of different levels of genetic structure. Twenty-two of 50 species of marine invertebrates show some level of significant structure along the North American west coast. Most structure occurs north of Monterey (based on R. Kelly and Palumbi, 2010). In this figure, PhiFT is a measure of genetic differentiation among populations. > High res figure
|
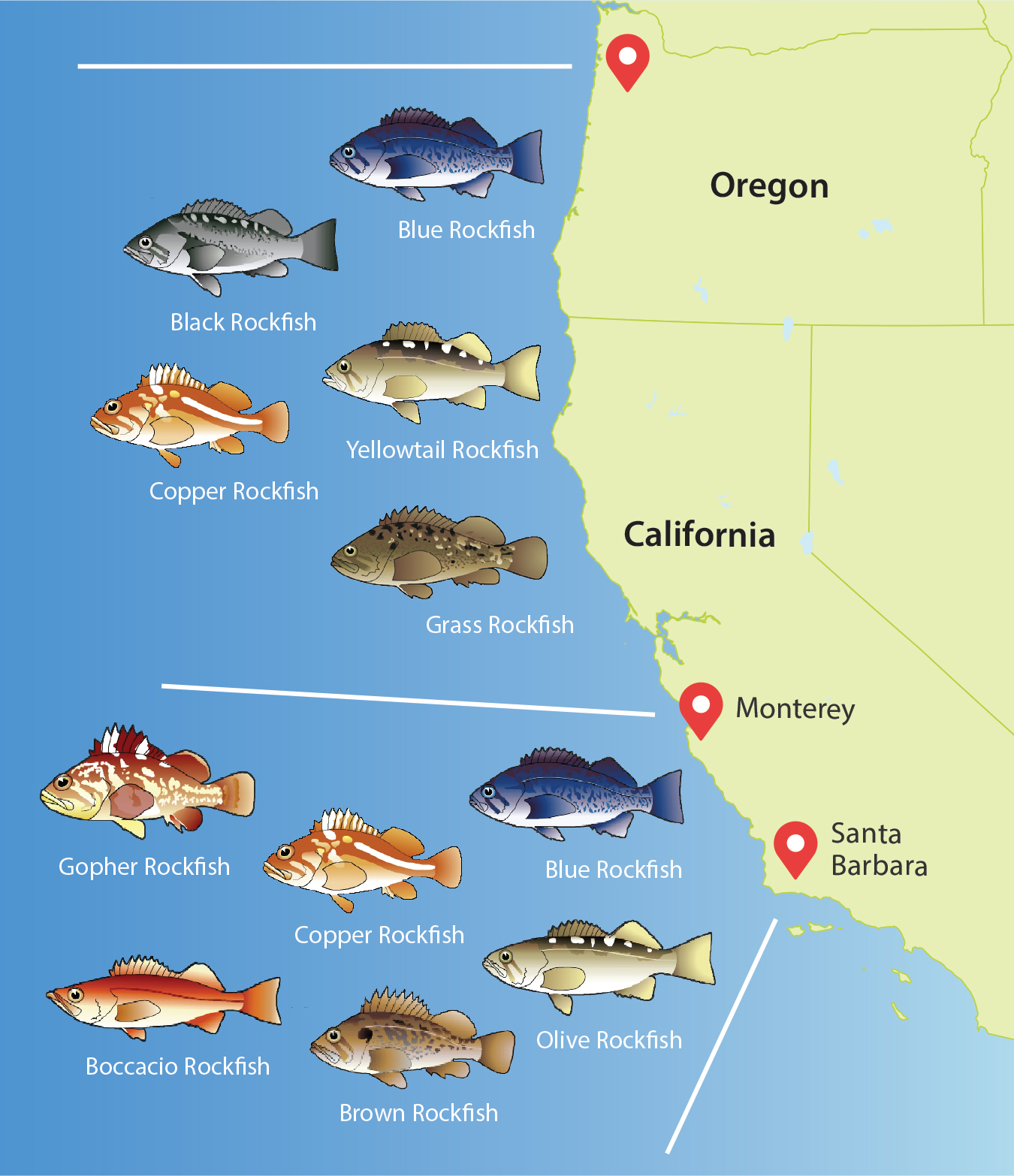
FIGURE 6. Nine of 11 rockfish species show strong West Coast differentiation, either between Oregon and Monterey or Monterey to Santa Barbara. Data from Sivasundar and Palumbi (2010); fish species artwork by Larry Allen and figure by M. Kovacs. > High res figure
|
These studies summarize strong structure over large geographic distances based around major oceanic features. However, numerous studies suggest that marine population structure can also sometimes be discovered across smaller spatial scales. The best characterized is a strong cline in genetics of the intertidal barnacle Balanus glandula, in which populations shift from northern genotypes at many loci to southern genotypes along the coast of central California (Sotka et al., 2004; Galindo et al., 2010). This species also shows genetic structure in central Oregon that depends on the strength of local upwelling (Barshis et al., 2011). Strong differentiation over short spatial scales from southern Oregon to northern California also occurs in blue rockfish (Sebastes mystinus). In this case, the genetic distinctions are strong enough to suggest that these different lineages may be different cryptic species (Burford, 2009). Even more genetic mosaicism is seen in low dispersal species such as the giant kelp (Macrocystis pyrifera; Johansson et al., 2015). In this species, fine-scale genetic differences have been recorded along Point Conception, California, and along the coast and among the islands of the Santa Barbara Channel.
In addition to the role of strong natural selection in the Levene (1953) Model, Richardson et al. (2014) also list other factors that might spark local adaptation over small spatial scales: seascape barriers, spatially autocorrelated environmental variation, and habitat selection. These features are likely contributors to the adaptive patterns outlined here as well as in other large marine ecosystems. For example, seascape barriers such as capes and promontories are often associated with genetic differentiation (see Figure 6), and marine ecosystems that stretch across latitudinal gradients have many spatially correlated environmental traits. Habitat selection plays a role in settlement in most marine species, and could alter gene flow across microhabitats. These features change the population and evolutionary setting for local adaptation and could make it a more common occurrence in ocean settings.
A NEW GENETIC FRONTIER—ASSEMBLAGE-WIDE ASSESSMENT OF DIVERSITY AND VARIATION BASED ON COMMUNITY DNA
In addition to the evolutionarily oriented studies detailed above, new genomics tools have recently become valuable as a way to complement and expand traditional ecological surveys of complex communities. Monitoring community diversity through long-term studies of species abundance and distribution (e.g., Blanchette et al., 2008) has long been a fundamental goal of PISCO and other large-scale ecosystem surveys. Over the 20 years of this interdisciplinary work, new tools have emerged that use DNA variation across species to help conduct monitoring of benthic species from collections of whole communities (community DNA) or even seawater (environmental DNA; Shum et al., in press). A key advantage of this approach is that it scales up more efficiently than do diver-focused measurements in kelp forests or even intertidal quadrat or transect surveys. This is likely to allow broader surveys of large-scale ecosystem dynamics along the North American west coast and elsewhere.
Recently, we have been testing community DNA surveys from kelp forest benthic samples as a way to monitor invertebrates and algae (Shum et al., in press). For example, we obtained DNA mixtures from the animals and algae on 97 kelp forest cobbles. From cytochrome oxidase gene amplifications, we recorded 406 species with good DNA identifications (>95% match to taxonomic data bases) from 14 animal phyla and three major algal groups (red, green, brown algae). These include many species of small amphipods, copepods, gastropods, and polychaete worms as well as large species such as eight common echinoderms. The wide diversity of species monitored and the relative ease of scaling up sampling locations make these cobble surveys an attractive addition to standard methods.
For these habitats, we can compare ecological and molecular surveys. PISCO survey transect data record 58 species, with the most records for large echinoderms and algae. The two data sets match reasonably well for echinoderms (Figure 7a) though there is high variance for rare species. Gastropods also have good correspondence, though there are many more small and hidden species—species likely to be missed by field observers—in the DNA data set. Arthropods do not match well between data sets—possibly because large mobile crabs (the dominant crustacean in the PISCO field surveys) are not collected on cobbles, and small cryptic isopods, amphipods, and copepods are not targeted in PISCO counts.
One method for reducing the biases in field survey data sets is to base the species abundance data on a presence/absence matrix instead of using the absolute number of individuals counted or the number of DNA sequences produced. For example, the most abundant species among our 97 cobbles are likely those that occur on more of them, so such presence/absence metabarcoding (PAM) can bring the DNA data more in line with survey data (Figure 7b).
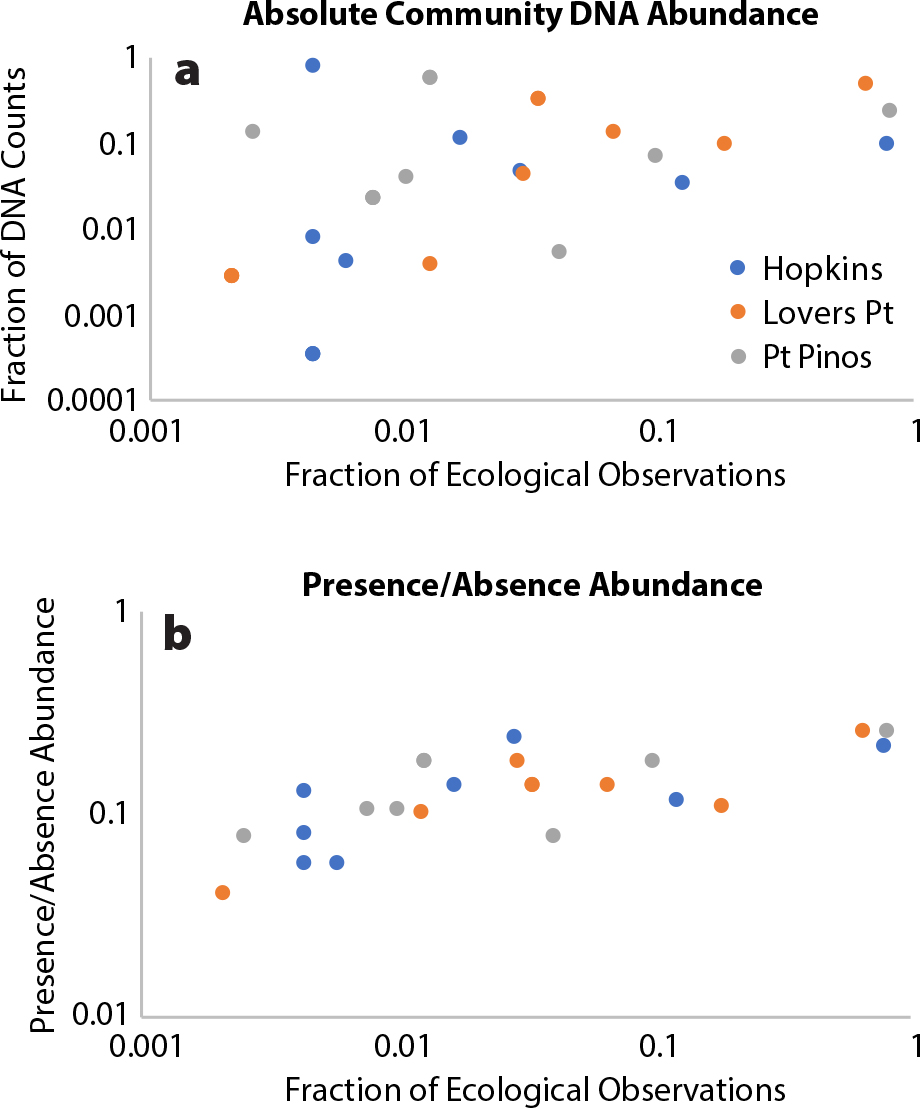
FIGURE 7. Echinoderm comparisons between PISCO field surveys and community DNA data sets for abundant echinoderm species. PISCO data are the number of counts of each of seven species at three locations in Monterey Bay (Hopkins Marine Station, Lovers Point and Point Piños) divided by the total number of those species from subtidal surveys. The DNA data come from mixtures of DNA from 97 cobbles collected from those three locations. DNA data derive from: (a) the number of DNA sequences in the combined DNA mixtures from each species, and (b) an assay of species abundance from the presence/absence matrix of each species on each cobble. The presence/absence data set reduces the variance at low abundance levels. > High res figure
|
Community DNA approaches scale up more easily because large numbers of samples can be centrally processed and effectively compared. Advantages of DNA surveys include that they require less field time; they can be focused on many more species, especially small invertebrates and cryptic algae; and they can provide additional data on microbes, disease, and other aspects of community dynamics. Yet, they still require careful fieldwork for collecting and handling specimens, and are unlikely to completely replace diver-led surveys. The ability to use these new tools for more comprehensive biotic surveys, for early warning of invasive species, and for surveys of settlement and ecological co-occurrence suggests joint development of community DNA and traditional ecological surveys.
IMPLICATIONS FOR CLIMATE RESPONSE
One of the summary messages from this compilation of PISCO-supported research is that marine species along the North American west coast are replete with genetic diversity. Moreover, genetic variation that appears adaptive at the level of gene function, expression, or diversity appears commonly in experiments using temperature or acidification. It may be that the highly variable environments of this coastline, the distribution of variation over time and space, and the large population sizes and high dispersal ability of many marine species have led to the evolution of adaptive genetic variation in many species. Strong selection may help tune populations for better performance in local environmental conditions.
This suggests that population adaptation to future conditions is possible and could potentially blunt the negative impact of climate change on these populations. This would be good news for the future of these populations. However, at least four critical questions remain unanswered about this possibility. First, the limits of this adaptation are not clear. Single experiments successfully show adaptive shifts, but what are the limits of this adaptation? How much adaptive potential is there? Second, these adaptive changes nearly all seem to be associated with trade-offs—in, for example, energy costs and metabolic flexibility. Such trade-offs are, in fact, predicted by models of spatial balancing selection, and they may limit future adaptation. Third, most experiments have studied gene function and adaptation to a single environmental stressor. But climate change will arrive with warmer, more acidic, more hypoxic water—is there genetic variation in most populations that could respond to all these variables simultaneously, and are genetic adaptations to them positively, negatively, or non-correlated?
Lastly, the adaptive mechanisms we can observe may result in evolution toward climate-adapted populations (e.g., Catullo et al., 2015), but at what rate? Climates are changing faster now than in the past, and it is not clear that the adaptive genetic variation evolved in the past will fuel the rapid adaptive evolution marine species need to keep pace with ongoing change (van Oppen et al., 2015). Important areas for future research include understanding if the genetic bases of locally adapted traits are compatible and whether dispersal and the spread of adaptive alleles up and down the coast will be fast enough to keep pace with the rates of environmental change.
Acknowledgments
The authors thank Kristen Milligan for her long-standing administrative and creative thoroughness in all things PISCO. Funding came from PISCO as well as the National Science Foundation, the Gordon and Betty Moore Foundation, and the David and Lucile Packard Foundation through the Hopkins Marine Life Observatory and PISCO. We also thank Moni Kovacs for her work on Figures 3,4, and 6.









