INTRODUCTION
One of the most damaging environmental catastrophes of our generation, the Deepwater Horizon (DWH) oil spill discharged 4.9 million barrels of oil and 250,000 metric tonnes of natural gas into the Gulf of Mexico and contaminated vast areas of the open ocean, the deep sea, and the shoreline. Its disruption of ecosystems carries residual and long-lasting impacts.
At the core of all ecosystems are microbial communities that constitute the foundation for all life by performing essential services such as carbon and nutrient cycling, as well as other core biogeochemical processes. Microbes are Earth’s first responders, and by rapidly reacting and adapting to changing conditions, they restore balance and stability to the entire ecosystem. In the context of oil spills, microbes serve as emergency cleanup crews by feeding on hydrocarbons and contributing other fundamental remediation services.
Despite their importance, relatively little was known about hydrocarbon-degrading marine microbes resident in the Gulf of Mexico prior to the DWH spill or about the effects of hydrocarbons on the microbiology of the Gulf region. The Gulf of Mexico Research Initiative (GoMRI) was established to provide resources for advancing understanding of marine hydrocarbon microbiology among other Gulf biological, physical, and chemical components. The microbiology research included development and application of genomic and bioinformatics tools that enabled scientists to examine hydrocarbon-degrading microbes in the context of complex microbial communities and in unprecedented detail. With an array of transformational “omics” tools, scientists gained valuable insight into how microbes respond to hydrocarbon infusions and restore ecosystem health. This research discovered novel species, genes, metabolic pathways, and community dynamics that are instrumental to hydrocarbon degradation and related ecosystem functions. In combination, the findings revealed an extensive and intricate natural capacity of microbes in the Gulf of Mexico to catalyze bioremediation of petroleum hydrocarbons. This new information will guide future mitigation and restoration strategies to harness the natural bioremediation capabilities of microorganisms without further disturbing sensitive ecosystems.
Broadly, the GoMRI researchers have provided an outline of core ecological and evolutionary principles regarding microbial response and ecosystem functions that is widely translatable to other systems. Lessons learned from GoMRI research form a foundation for understanding how microbes in various ecosystems around the globe respond to diverse environmental disturbances. From the Arctic to the equator and the deep sea to coastal shores, these principles apply to describing how microbes maintain and restore ecosystem balance. These insights will help scientists better understand and prepare for future catastrophes, from a tanker spill to the long-term disruptions of climate change.
In summary, the research carried out by GoMRI investigators led to new methods for monitoring and assessing ecosystem health using microbial populations as ecosystem indicators in marine and coastal environments. Scientists can now deploy omics tools to take the pulses of microbial communities, identify disturbances, and guide mitigation strategies—notably at early stages before widespread damage occurs. Although much work is still needed to fully understand and engage microbially mediated biogeochemical processes that underpin ocean systems and their resilience, GoMRI research has prepared future generations to better protect and restore invaluable habitats by guiding disaster preparedness and response in the face of diverse environmental stressors and natural disasters around the globe.
OMICS TECHNOLOGIES ENABLE NEW BIOGEOCHEMICAL DISCOVERIES
Discovery of New Oil-Degrading Microbial Species, Genes, and Enzymes
A powerful toolbox of omics technologies and bioinformatic methods, including genomics, transcriptomics, proteomics, metabolomics, and metagenomics, is now available that allows scientists to probe the structures and functions of microbial communities that form the foundation of marine ecosystems (Grossart et al., 2020). These technologies provide researchers with the tools to comprehensively examine microbial dynamics in real-world situations (Mason et al., 2014a,b; Rodriguez-R et al., 2015). The advanced techniques and bioinformatics methods used generated discoveries and impacts far beyond what was previously thought possible (Karthikeyan et al., 2019).
GoMRI researchers discovered many new microbial taxa that thrive in oil-contaminated marine environments, along with novel genes and metabolic pathways they use for hydrocarbon degradation (see image on opening page of this article; Hazen et al., 2010; Kostka et al., 2011; Mason et al., 2014a; Crespo-Medina et al., 2014; Rodriguez-R et al., 2015; Kleindienst et al., 2015a; Yang et al., 2016; Karthikeyan et al., 2019). In addition, major known classes of microbes, for example, Bacteroidetes, were found to harbor a previously unrecognized potential for hydrocarbon biodegradation (Liu and Liu, 2013). However, the many major players and ecotypes discovered represent only a fraction of all the microbes involved in hydrocarbon degradation (Gutierrez et al., 2018).
Oil-Degrading Microbial Collaboration and Community Dynamics
Oil-degrading microbes are ubiquitous in marine and estuarine environments around the world, but their abundance is low when oil is absent. As part of the “rare biosphere,” they harbor metabolic potential of ecological significance; they act as first responders in the event of an oil spill, providing critical ecosystem cleanup and stabilization functions that impact the fate and environmental distribution of hydrocarbons.
When oil spills occur, microorganisms sense and respond to the oil, using innate mobility to pursue oil via flagella-based motility and chemotaxis. They are stimulated to express hydrocarbon degradation genes that enable metabolism of various hydrocarbon compounds as energy sources. These hydrocarbon utilizers reproduce rapidly in the presence of oil to dominate the microbial ecosystem in contaminated waters and sediments, creating a microbial bloom (Kostka et al., 2011; Doyle et al., 2018, 2020; Karthikeyan et al., 2019). Many oil-degrading microbes possess multiple pathways for hydrocarbon degradation, and which metabolic pathway is induced may depend on environmental conditions and type of exposure (Karthikeyan et al., 2020a).
Microbes often exhibit cooperative metabolism by partitioning metabolic pathways among community members (Zengler and Zaramela, 2018). These microbial partnerships or collaborations are critical to microbial community functions and processes, including hydrocarbon degradation (Joye et al., 2016). Microbes can also create new catabolic pathways by shuffling hydrocarbon degradation genes between and among community members via lateral gene transfer, generating novel combinations of enzymes and metabolic capabilities in new hosts.
GoMRI research led to the discovery of metabolic partnerships for oil degradation. Metagenomic analysis revealed that genes within a single metabolic pathway may be distributed throughout the community, rather than contained within a single microbial species or strain. In some cases, different bacterial species were found to specialize in specific metabolic steps, but because they shuttled metabolites among species, collaborative networks were created that together completed the oil degradation process. In other cases, researchers determined that certain species specialize in breaking down the byproducts that are generated by other species. Together, these microbial networks can accomplish far more, with greater efficiency, than any single species could. Taking such a holistic approach, the research done by the GoMRI scientists has illuminated collective metabolic pathways and ecosystem functions that would have been overlooked by examining only individual species (Mason et al., 2012; Doyle et al., 2020).
In short order, the DWH spill caused major shifts in microbial ecosystem structure and function. Rare biosphere species capable of degrading hydrocarbons rapidly outgrew neighbors, ultimately accounting for up to 90% of the community (Kleindienst et al., 2015a; Karthikeyan et al., 2019, 2020a). At the same time, microbes that typically dominate healthy environmental systems, such as ammonia oxidizers and other autotrophs and heterotrophs, declined in relative abundance. Coastal nitrifier populations in offshore pelagic waters underwent alteration in species composition for at least a year, returning to normal only after the oil had dissipated. (Newell et al., 2014; Huettel et al., 2018)
Community composition shifted over time as hydrocarbon oxidizing bacterial blooms consumed available hydrocarbons, and then declined, being replaced by microbial species capable of degrading the residual metabolic byproducts (Figure 1; Kostka et al., 2011; Rodriguez-R et al., 2015; Kamalanathan et al., 2020). Such microbial succession patterns have been documented across many different ecosystems, each with unique species and dynamics, driven by abundance of different hydrocarbon fractions and related environmental factors and at different time points. Thus, conclusions can be drawn from integrating data across time and localities but must be contextualized when making broader observations (Crespo-Medina et al., 2014).
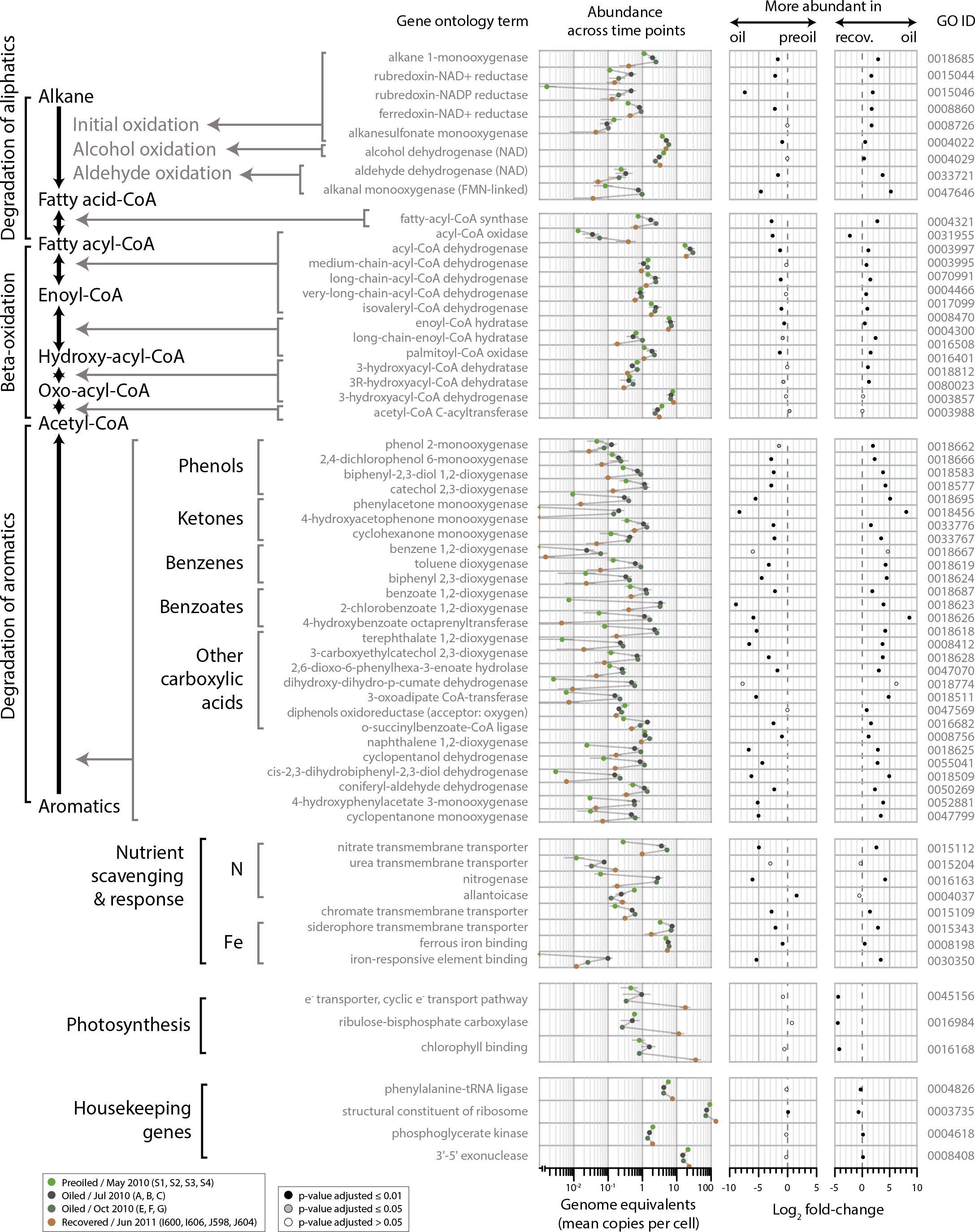
Figure 1. Microbial community functional shifts in coastal sediments in response to oil from the Deepwater Horizon discharge. Genes coding for selected molecular functions related to hydrocarbon degradation, nutrient scavenging and response, photosynthesis, and housekeeping genes are listed (left), with mean genome equivalents per group of samples (middle), and log2 of pre-oil/oiled and oiled/recovered (recov.) fold changes (right). Gene abundance was assessed as the average genome equivalents (mean copies per bacterial cell) at each sampling period. The log2 fold change was estimated as the log2 of the ratio of normalized counts between pre-oiled samples (S1 to S4) and oiled samples (A to G) and between oiled samples and recovered samples (I600, I606, J598, and J604). P values were estimated using a negative binomial test. Interesting patterns were the succession of genes related to easily degradable, light-hydrocarbon fractions, followed by genes specializing in polycyclic aromatic hydrocarbons (PAHs) and complex aromatics, and an increase in nitrogen fixation genes that denote nitrogen limitation for the microbial communities during crude oil biodegradation. CoA = coenzyme A. Figure from Rodriguez-R et al. (2015). > High res figure
|
In the deep-sea plume, methane, and potentially propane, butane, and ethane, fueled the early microbial response (Valentine et al., 2010; Joye et al., 2011; Crespo-Medina et al., 2014). Methane was a major driver in the DWH spill, spawning the emergence of Methylomonas species with methane oxidation capabilities (Figure 2; Crespo-Medina et al., 2014). Although Archaea are known generally to play important roles in anaerobic methane oxidation and anaerobic hydrocarbon (oil) degradation, they were not major players in pelagic or benthic oil or gas degradation during DWH. Immediately after the spill, when n-alkanes and cycloalkanes were more abundant, Oceanospirillaceae and Pseudomonas spp. were dominant (Figure 3; Hazen et al., 2010). These species were later supplanted mainly by Colwellia spp. and, to a lesser degree, Cycloclasticus and Pseudoalteromonas spp., which peaked when linear and simple aromatic hydrocarbons were abundant (Dubinsky et al., 2013; Kleindienst et al., 2015a). Employing metagenomics, scientists confirmed that the different hydrocarbon degradation genes and pathways present at different time points corresponded with abundances of different substrates within the plume (Mason et al., 2012; Redmond and Valentine, 2012).
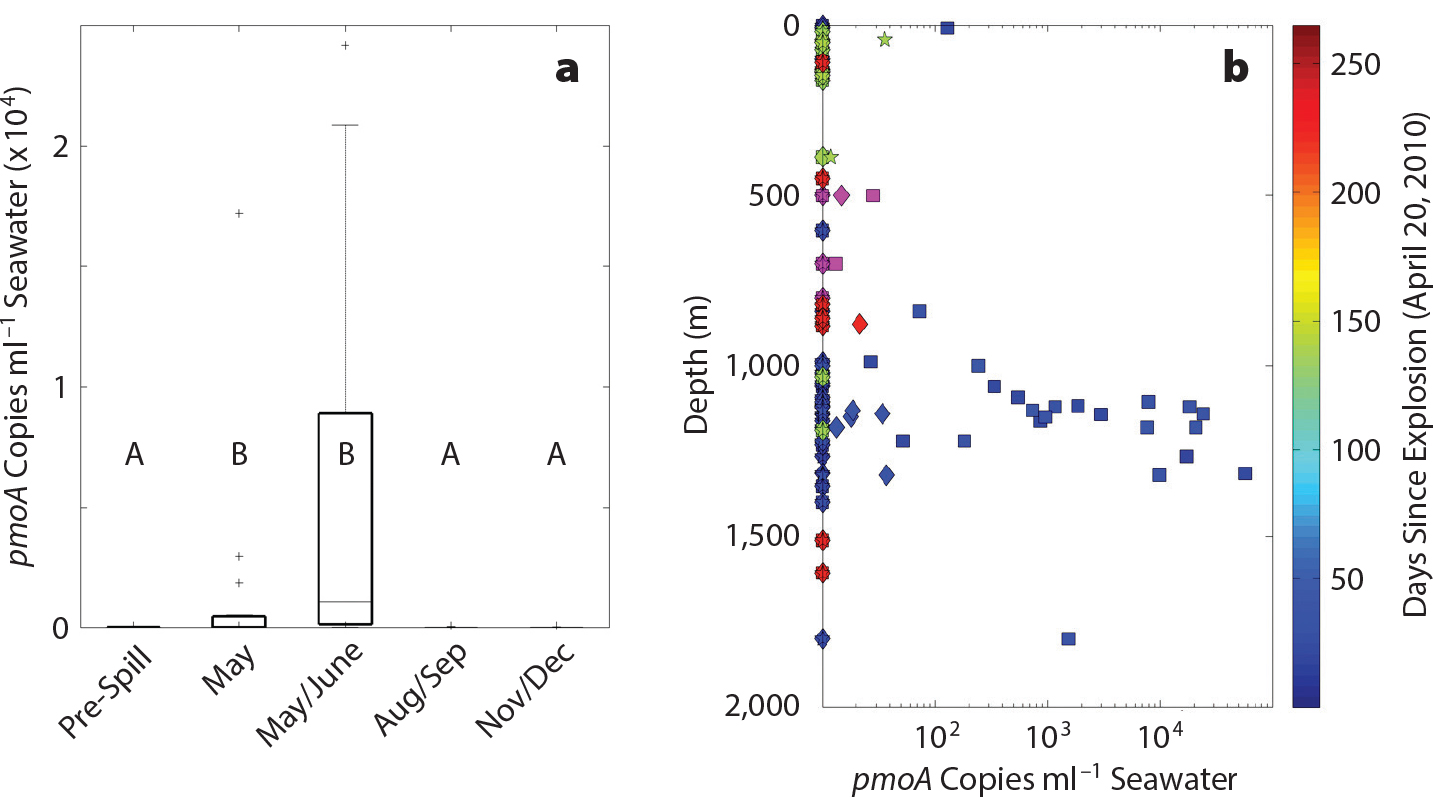
Figure 2. Abundance of pmoA genes following the Deepwater Horizon discharge. (a) Abundance of methanotrophic bacterial pmoA genes. Data are binned by time period. (b) Abundance of pmoA gene copies over time (pre-spill samples are magenta). Stars = OPU1. Diamonds = OPU3. Squares = New phenotype. Data in the two time periods marked with asterisks in panel (a) are different from those collected at other times, with a statistical significance of a P value of <0.05. Plus signs denote extreme data outliers. Figure from Crespo-Medina et al. (2014) > High res figure
|
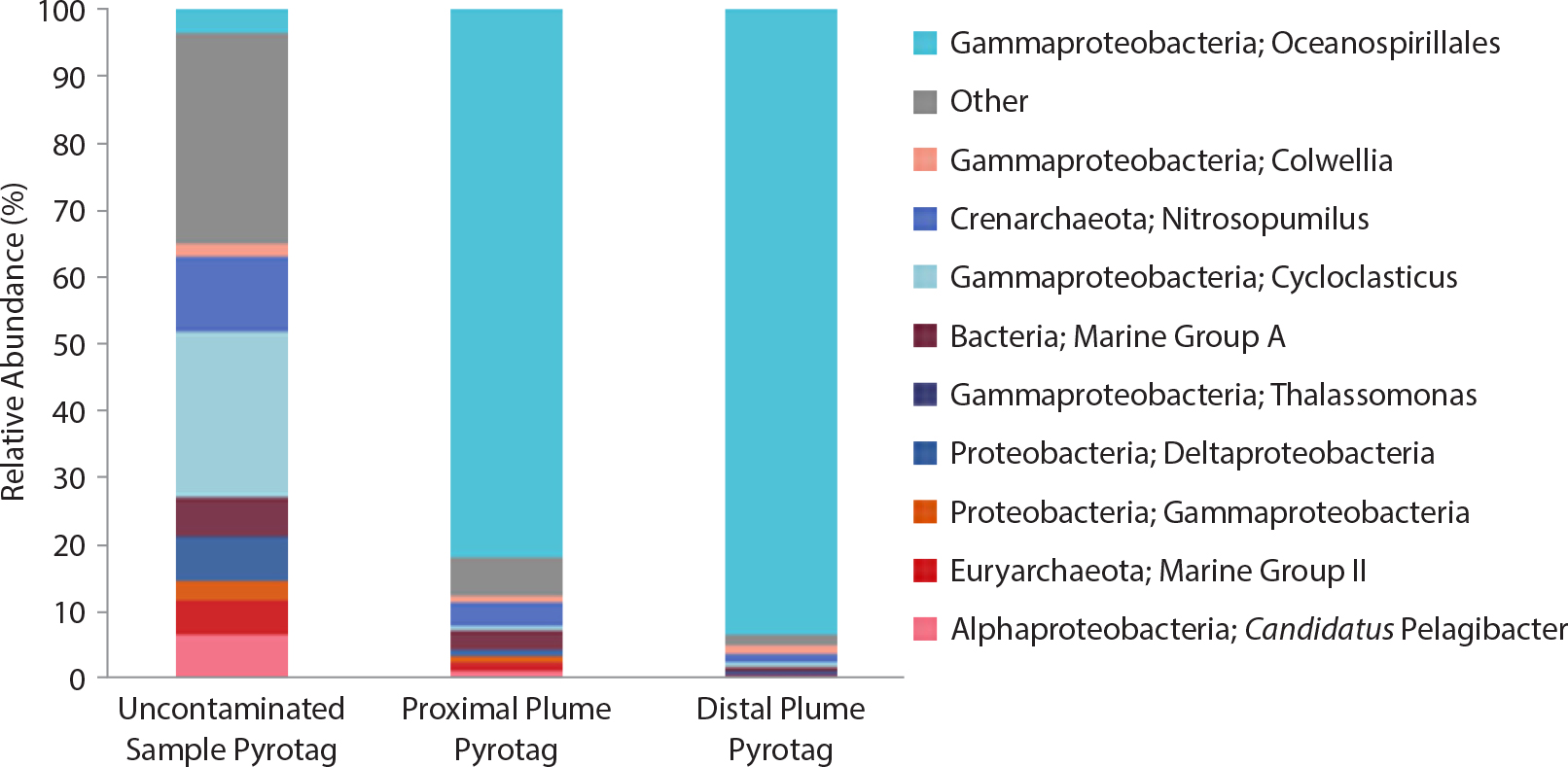
Figure 3. Microbial community structure and function change drastically in response to hydrocarbons. Community structure, function, and succession patterns varied depending on location and time of sampling. Figure modified from Mason et al. (2012) > High res figure
|
Meanwhile, in beach sands, succession of microbial populations also paralleled the chemical evolution of petroleum hydrocarbons (Rodriguez-R et al., 2015). Early responders were mostly Gammaproteobacteria (Alcanivorax and Marinobacter spp.), microbes known to degrade aliphatic hydrocarbons, and their bloom coincided with a drastic decline in community diversity (Kostka et al., 2011). These were replaced after three months by Alphaproteobacteria (Hyphomonas and Parvibaculum spp.), capable of aromatic hydrocarbon decomposition. This shift coincided with the disappearance of alkanes and persistence of aromatics in field samples (Huettel et al., 2018). Nearly all of the oil on Pensacola Beach was degraded after one year, and microbial communities returned to a state that resembled the pre-oil condition, with nearly all known hydrocarbon degraders and genes coding for degradation functions exhibiting only low or undetectable levels (Rodriguez-R et al., 2015). However, significant differences were observed between the pre-oiled and recovered communities. Taxonomic diversity remained elevated in recovered communities, above that of the pre-oiled community, while functional diversity was lower in the recovered compared to the pre-oiled. Such differences could be due to unrecognized long-term effects of disturbance, such as the emergence of new taxa, and stochastic environmental events such as changes in nutrient or carbon input (Kostka et al., 2020). This return to a “baseline” state remains an open question that is actively being pursued as a legacy DWH issue.
In salt marshes, the microbial diversity of aerobic and anaerobic sediments decreased, favoring microbes capable of degrading alkanes and aromatics as weathered oil and residues accumulated (Atlas et al., 2015). In anoxic sediments, sulfate-reducing bacteria, for example, Desulfococcus spp., increased in parallel with oil contamination. Interestingly, marsh sediments maintained a high percentage of hydrocarbon degraders whose growth was driven periodically by storm surges that redistributed weathered oil from the anoxic sediment. These lingering shifts in microbial community diversity and function have the potential to impact the overall marsh ecosystem, particularly marsh vegetation and fishery health.
In anoxic environments (e.g., the seafloor and salt marsh sediments), biodegradation occurs much more slowly so that most of the oil persists. (Liu et al., 2012; Chanton et al., 2015). Yet, seafloor sediments near the DWH wellhead showed some enrichment, notably in anaerobic respiration and anaerobic hydrocarbon degradation genes and associated species such as Deltaproteobacteria (Mason et al., 2014b). Novel anaerobic polycyclic aromatic hydrocarbon (PAH) degradation pathways were also discovered (Shin et al., 2019b), potentially mediated by yet undiscovered sulfate-reducing bacteria along with their fermentative syntrophic partners. Hexadecane-degrading microbes, closely related to the Desulfobacteraceae, and phenanthrene-degrading microbes related to Desulfatiglans spp. were also discovered.
Biogeochemical Processes
Microbes play fundamental roles in basic biogeochemical cycles of marine and coastal ecosystems, including interconnected carbon, nitrogen, and phosphorus cycling—and also in hydrocarbon degradation. Omics analyses of these pathways revealed how these ecosystem functions were impacted by the DWH spill.
Nitrogen Fixation
Like all microbes, oil degraders require substantial amounts of nitrogen for growth, but nitrogen is present only in limited amounts in marine environments. Although nitrogen fertilizers have been added to accelerate microbial growth and biodegradation following some oil spills, such as the Exxon Valdez spill in Prince William Sound, Alaska (Pritchard et al., 1992), microbes can generate their own fixed nitrogen through biochemical fixation of dinitrogen gas (Foght, 2010). Nitrogen fixation pathways are well known in soil microbes that support crop growth and in oceanic microbes such as Trichodesmium cyanobacteria (Bergman et al., 2013), but nitrogen fixation by oil degraders in response to raw oil exposure is a new discovery (Quigg et al., 2016).
Researchers studying the DWH spill discovered that nitrogen fixation is a key ecosystem function of microbial communities in response to oil (Rodriguez-R et al., 2015; Fernández-Carrera et al., 2016). In the offshore and coastal ocean ecosystems where nitrogen is limiting, nitrogen fixation function increases in response to oil contamination (Scott et al., 2014; Rodriguez-R et al., 2015; Gaby et al., 2018; Shin et al., 2019a), and then dissipates once the hydrocarbons are significantly reduced (Shin et al., 2019a).
Fixing nitrogen provides a strong selective advantage for those species capable of fixation in the presence of oil, allowing them to utilize hydrocarbon energy sources without restriction. Although species with such dual capabilities are uncommon under pristine conditions, they thrive in oil-contaminated environments. This explains the niche dominance of keystone oil-degrading organisms such as Candidatus Macondimonas diazotrophica (Karthikeyan et al., 2019).
Microbial genes involved in carbon, nitrogen, phosphorus, sulfur, and iron cycling were also enriched in oil-contaminated ecosystems (for additional information, see Mason et al., 2012; Rodriguez-R et al., 2015). Understanding these biogeochemical interdependencies could inform fertilization strategies to enhance natural biodegradation.
Marine Oil Snow (MOS)
Phytoplankton and hydrocarbon-degrading microbes that assemble around oil droplets in large communities produce transparent exopolymers (TEPs) that entrap oil. These dense macroscopic assemblages of carbohydrates and biomass behave like “snow” as they sink to the seafloor. Marine oil snow (MOS) effectively transfers hydrocarbons from the water column to the seafloor as sediment (Vonk et al., 2015). Although MOS formation was observed during previous oil spills, including the 1977 Baltic Sea Tsesis spill (Johansson et al., 1980) and the 1979 Ixtoc I spill in the southern Gulf of Mexico (Patton et al., 1981), it was first thoroughly studied after the DWH spill, which triggered MOS formation in unprecedented quantities (Joye et al., 2014).
Oil degradation and deposition that takes places in MOS impacts the fate of spilled oil (Figure 4). MOS particles are hot spots for oil degradation, exhibiting high levels of lipase activity (Gutierrez et al., 2018) and distinct microbial communities that specialize in breaking down oil. In particular, Colwellia, Marinobacter spp., and Alteromonas spp. are prevalent because they have the unique capacity to degrade oil relatively rapidly in cold, deep marine environments (Gutierrez et al., 2018).
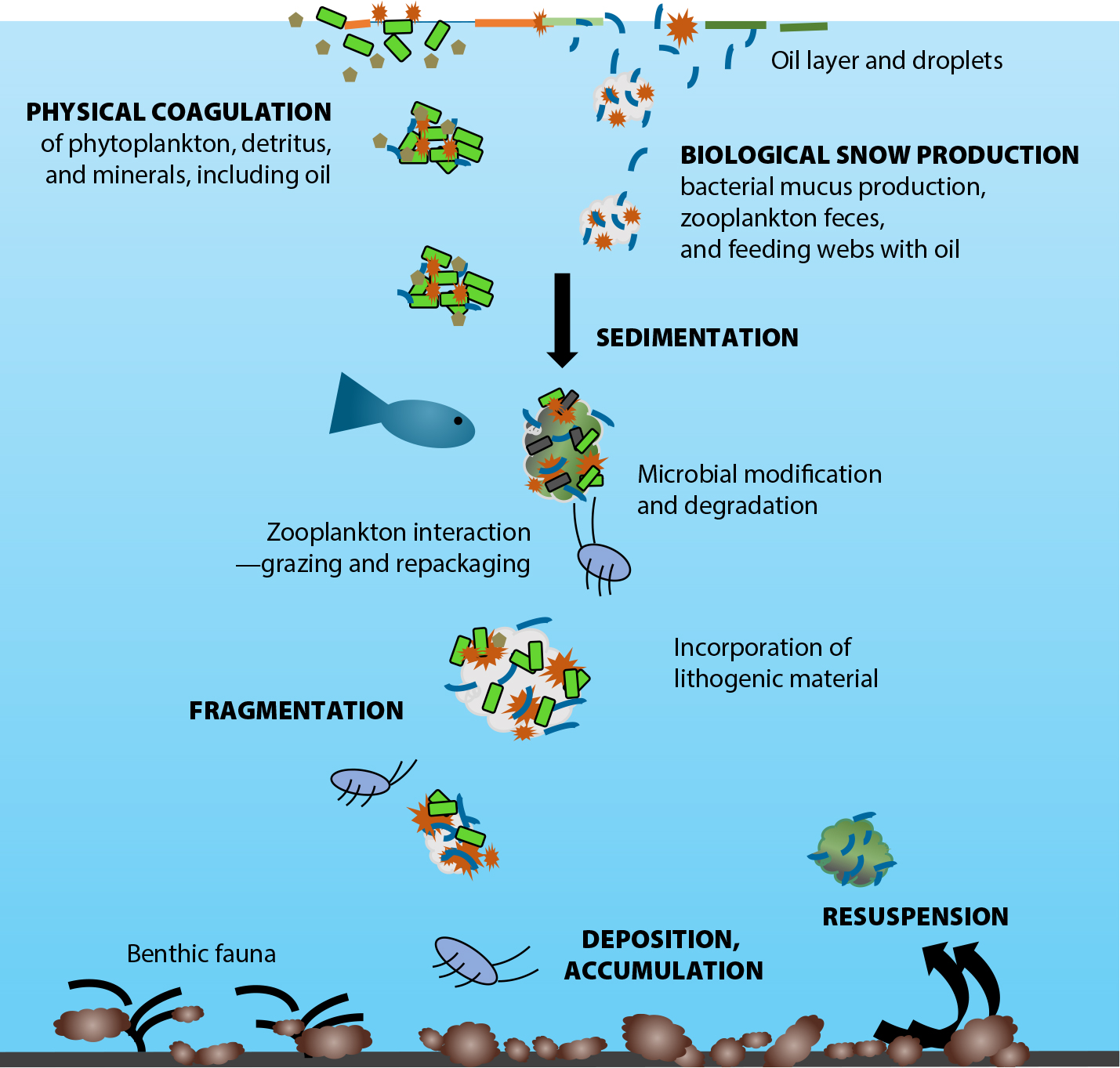
Figure 4. The formation, sinking and loss of marine oil snow, which consists of exudates, biotic, and abiotic particles. Marine oil snow is a common food source that removes hydrocarbons from the water column as it sinks to the seafloor. Image credit: Uta Passow, Memorial University of Newfoundland. > High res figure
|
MOS particles sink quickly, reaching the seafloor within ~10 days (Passow, 2016). MOS formation is triggered by water column elements that include detritus, fecal matter, minerals, and chemical dispersants, as well as such living cells as diatoms, microalgae, and bacteria, and the polymers they secrete.
The question remains, however, whether MOS is good or bad as an oil spill bioremediation mechanism. MOS could have negative consequences for the water column as well as the seafloor where it is ultimately deposited. These larger particles can trap and remove planktonic animals from the water column. MOS particles can also effectively seal off the sediment surface, potentially suffocating fauna, while addition of organic content to the benthos could stimulate respiration, with negative consequences for filter-feeding fauna. Understanding the consequences of MOS in the water column and the benthos requires further investigation.
SIGNIFICANCE AND APPLICATIONS OF GoMRI RESEARCH
Ultimately, scientific and technological advances derived from GoMRI research provide a foundation for new research that will improve strategies for predicting and mitigating ecosystem perturbations and have impacts across broad scientific disciplines and far beyond the Gulf of Mexico.
Broad Lessons in Microbial Hydrocarbon Response
As the examples cited above show, GoMRI research has revealed ecological, evolutionary, and biogeochemical principles that underlie microbial community composition and response to disturbance across diverse environments and contexts.
The hydrocarbon-degrading microbes and genes that code for hydrocarbon degradation and utilization discovered by GoMRI scientists are present in oil-contaminated waters and coastal regions around the world, and thus are available everywhere as first responders. However, while oil-degrading microbes are globally distributed, regional variables can significantly influence microbial community composition and metabolic capabilities prior to and during a spill.
Physical, chemical, and biological environmental conditions, including temperature, salinity, hydrostatic pressure, oxygen supply, nutrient availability (particularly N and P), water currents and stratification, MOS presence, and sea ice can influence microbial species composition and rates of hydrocarbon degradation, activity, and dispersal from the deep sea to intertidal marsh ecosystems (Head et al., 2006; Edwards et al., 2011; Redmond and Valentine, 2012; Rodriguez-R et al., 2015; Fernández-Carrera et al., 2016; Huettel et al., 2018; Sun and Kostka, 2019). Furthermore, the type of oil spilled, how it weathers, and the extent of sunlight exposure can alter the chemical composition of oil and/or induce MOS formation, all of which act to drive different microbial responses (Bacosa et al., 2015; Shin et al., 2019a). The DWH spill provided a unique opportunity to study many of these environmental factors because it affected a wide variety of open-water and coastal ecosystems with large variations in physical and chemical conditions.
Microbial communities in the Gulf of Mexico are considered to have been uniquely primed for rapid response to oil spills due to the prevalence of natural hydrocarbon seeps and drilling operations that constantly discharge oil into the Gulf. The background levels of hydrocarbons sustain a higher proportion of oil-degrading microbes that are also typical of many other regions with natural seepage and/or hydrothermal venting, for example, the Gulf of California, the Bay of Bengal, the Black Sea, and the Arctic Ocean. In addition, shipping lanes in open waters and ports are primed for oil response because of long-term industrial and human activities. In some cases, hydrocarbons produced by phytoplankton or cyanobacteria can prime microbes for hydrocarbon degradation through low level exposure (Guiterrez et al., 2014; Love et al., 2021).
Oil spills are particularly threatening to Arctic regions where climate change is already wreaking havoc. It is therefore critical to understand the environmental risks and develop mitigation strategies before a major spill occurs in these delicate ecosystems. Interestingly, GoMRI research has proven pertinent to this region; preliminary results of these studies reveal microbial responses and community succession similar to those reported for the Arctic marine environment (Sun and Kostka, 2019). However, sea ice and extremely low temperatures can alter degradation properties of both oil and microbial communities, which may significantly impact biodegradation and need to be considered. The Canadian government is actively engaged with the GoMRI community to build on GoMRI discoveries, methodology, and technology in efforts to prepare for a spill in the Canadian Arctic.
Informing Oil Spill Response
GoMRI research provides a foundation of knowledge and tools that will enable scientists to play key roles in guiding future oil spill response and mitigation planning using data-driven strategies. Evidence-based interventions informed by evaluating ecosystem responses and recovery status will improve the success of future emergency responses by replacing previous trial-and-error approaches.
Specifically, by examining the presence of oil-degrading species, genes, and biogeochemical pathways, scientists can assess the natural bioremediation potential of a community, providing insight into whether and how its metabolic capabilities can be augmented. One example is addition of nitrogen fertilizers, which has been proposed by GoMRI and other investigators to accelerate microbial bioremediation (Edwards et al., 2011; Kleindienst et al., 2015b; Fernández-Carrera et al., 2016). Additionally, enhancement of microbes that produce exopolymeric substances, which emulsify oil and act like dispersants, could preclude addition of man-made dispersants such as Corexit (Ziervogel et al., 2019). Strategies to promote growth of native microbial species or utilization of genetically engineered microbes for natural surfactant production would provide responders with biodegradable, nontoxic surfactant alternatives. Natural microbial surfactants may also prove effective as nontoxic dispersants with fewer environmental concerns (Bacosa et al., 2018).
Mounting an effective microbial oil spill response will require analysis of microbial communities not only during a spill but also before and after. Documenting the “normal” or “baseline” state of an ecosystem prior to a crisis is essential for planning intelligent mitigation strategies aimed at restoring native microbial communities and their ecosystem functions (Joye, 2015). In addition, it can indicate whether an ecosystem is compromised by previous environmental stress and therefore in need of additional protections, for example, limiting oil drilling or tanker traffic (Sun and Kostka, 2019). After a spill, monitoring mitigation and restoration efforts should be undertaken to assess whether response actions work as predicted or if alternative plans should be implemented.
In summary, with application of omics tools, scientists can assist first responders in determining potential environmental impacts of a spill, as well as actions to be taken, on what time frames, and in which locations to minimize risks and damage.
Assessing Ecosystem Health in the Context of Environmental Disturbance
By developing basic principles of microbial ecosystem function and response along with techniques to study them, GoMRI scientists have provided a basis for understanding diverse marine and terrestrial ecosystems in order to address natural and man-made disasters, from chemical spills to algal blooms, and even gradual and long-term stressors, including climate change. Understanding microbial ecosystem dynamics and function in response to climate perturbations will be essential for predicting long-term impacts and for preparing mitigation plans.
Documentation of indigenous microbial communities in diverse marine and terrestrial ecosystems around the world is necessary for assessing ecosystem changes and restoration goals in the context of any perturbation. Baseline metagenomic analyses require sampling over vast timescales to account for daily, monthly, and seasonal community variations, as well as interannual and even decadal shifts. Such studies are underway to catalogue Earth’s microbiome (Gilbert et al., 2018). From deepwater benthic regions to coral reefs, beach sands, marshes, and associated terrestrial ecosystems (Magnuson, 1990, 1995), different ecosystems each have unique microbial baselines that require individual documentation so that significant or problematic changes can be detected and measured.
Microbial Indicators of Ecosystem Health and Disturbance
GoMRI researchers identified microbial indicators of ecosystem disturbance in the context of the DWH spill that will help scientists assess ecosystem health across various contexts.
Low microbial diversity can be a red flag, signaling ecosystem disruption and declining ocean health. A healthy microbial ecosystem is composed of a variety of species that provide diverse ecosystem functions and maintain system stability. Environmental disturbances such as the DWH spill disrupt the balance, favoring a limited number of species that is able to adapt to new conditions. These few species thrive over the others, often reflected in low overall species diversity. A community with low diversity may be more sensitive and less resilient to further disturbances.
The presence of a single species or genus in very high numbers indicates a recent ecosystem disruption, even if the original composition of the community is unknown. For example, during the DWH spill, rare species with hydrocarbon degradation potential and/or nitrogen fixation capabilities were poised to take advantage of new energy sources. Under novel conditions, these species outcompete their neighbors and can grow to one-third or more of
the population.
The emergence of different and novel genes, metabolic pathways, and ecosystem functions within microbial communities reflects adaptation to environmental change. When microbes respond to environmental disturbance, organisms may express different genes and metabolic pathways as they adapt to new sources of energy (hydrocarbons), nutrients, physical/chemical limitations, and other factors in order to take on novel and adaptive ecosystem functions.
These genetic and metabolic indicators of ecosystem disruption can also provide critical clues as to how the environment has changed, and therefore how it might be restored.
“Using the powerful and constantly evolving tools of genomics, GoMRI researchers discovered novel [microbial] genes, pathways, organisms, communities, and partnerships associated with oil decomposition and spill remediation.”
|
THE FUTURE OF GENOMICS IN THE STUDY AND MANAGEMENT OF ECOSYSTEM HEALTH
In the future, scientists hope to identify microbial “biomarkers” of ocean health, as detectors of ecosystem disruption and risk predictors in the face of diverse threats, including man-made and natural disasters as well as climate change. For example, how might microbiologists help fisheries or oyster farms recover after an oil spill or hurricane? Similar to ways that medical biomarkers help doctors diagnose diseases and identify appropriate treatments, researchers could “take the pulse” of the ecosystem with microbial biomarkers to identify atypical functioning and direct strategies to correct these problems.
Biomarkers might be used in various ways to assess and support ecosystem health:
Predictive biomarkers. Disruptions in microbial communities can often be perceived before ecological consequences or impacts are apparent. Thus, microbes can serve as the “canary in the coal mine” to warn of impending consequences of a disturbance. For example, microbial community restructuring or functional changes might indicate a slow oil leak or gradual warming due to climate change. This can trigger early action to better prevent or mitigate potential damages from natural and man-made disasters in the ocean, as well as other ecosystems, by alerting scientists to changes before they are irreversible, and/or identifying areas that need extra protections.
Diagnostic biomarkers. The presence of specific indicator species and genes might be used to “diagnose” certain disturbances, which could indicate what has gone awry within an ecosystem. For example, overgrowth of a species and emergence of novel genes, metabolic pathways, and ecosystem function can all assist scientists in identifying biogeochemical disruptions.
Therapeutic biomarkers. Indicator species and genes might also point scientists and managers toward solutions by providing information about how an environment has changed and, therefore, how its ecosystem might be restored. Therefore, in addition to informing mitigation and restoration strategies, these indicators can help to identify action plans during a crisis event. For example, the emergence of a nitrogen-fixing species might indicate nitrogen as a limiting factor in bioremediation, suggesting that the addition of nitrogen fertilizers would accelerate recovery.
To assemble a complete picture of ecosystem health will require the use of multiple, diverse biomarkers, each of which will provide a piece of the puzzle. For example, genomic biomarkers may provide insight into hydrocarbon degradation, toxin accumulation, hypoxic conditions, or other environmental disturbances. Meanwhile, metatranscriptomics might reveal immediate adaptations, whereas meta-community restructuring suggests long-term consequences.
Ideally, such biomarkers will be able to inform scientists and first responders on site and in real time as an essential component of an environmental emergency response toolkit. This will require robust, portable (i.e., hand-held devices), cost-
effective, and integrative analytic instruments, such as the small mobile DNA sequencers designed for the rigors of field research.
Predicting Environmental Impacts with Biomarkers and Models
Biomarkers can be paired with biogeochemical models to predict important outcomes, including when oil will be removed from a system, how long would it take for an ecosystem to recover, whether responders should intervene to speed ecosystem recovery, and other aspects of biogeochemical responses and timelines. Models can also reveal which factors may have the greatest impact on damage or recovery, enabling responders to tailor mitigation strategies rather than relying on one-size-fits-all approaches that may or may not work.
Biogeochemical models can help scientists answer questions, including:
- Should oil spills be seeded with microbes with hydrocarbon degradation capabilities? If so, which ones and in what quantity?
- Should dispersants and/or fertilizers be added? If so, in what quantity? How will such interventions impact ability of indigenous microbes to degrade oil?
- What environmental impact can be avoided with an intervention? What are potential unintended consequences of intervention?
- Will leveraging natural microbial processes be more successful than alternative approaches such as burning or vacuuming the oil?
Current biogeochemical models are limited and, in general, unable to answer such complex questions. To achieve more realistic models will require significant advances in understanding physical, chemical, and biological processes and their interactions in the ocean, as well as active and effective collaborations among oceanographers, microbiologists, computational scientists, and many other disciplines.
The greatest hurdle in developing predictive biogeochemical models is translating omics data to metabolic functions. While metagenomics informs metabolic potential, it may not always reflect microbial activity. There are many novel microbial species, genes, and proteins to be discovered and identified, and their biogeochemical functions characterized, most probably with inferences from taxonomic and sequence similarities to reference species and their metabolic profiles. In reality, metabolic capabilities can differ dramatically from predicted activities and currently must be defined through cultivation, biochemical assays, or gene knockout experiments. Machine learning/artificial intelligence is likely to be of great value in overcoming this obstacle.
To better link omics data with functional rates of activity at the enzymatic and cellular levels will require new strategies, such as stable isotope probing (SIP) and biorthogonal noncanonical amino acid tagging (BONCAT). These tools track metabolic activity of individual microbes and consortia within their natural environments. They can assign specific paths to specific microbes, elucidate cross-feeding partnerships, and identify novel metabolic steps and pathways.
Facilitating Advances Through Open-Access Resources and Multidisciplinary Collaboration
The omics and environmental data sets collected by GoMRI investigators are accessible via the open-access platform https://data.gulfresearchinitiative.org/. In the event of a new spill or disaster, this resource can be mined to predict consequences and inform response.
Specifically, GoMRI researchers created the Genome Repository of Oil Systems (Karthikeyan et al., 2020b), a comprehensive, searchable database that documents microbial populations in natural oil ecosystems and oil spills, along with available underlying physicochemical data, geocoded via the geographic information system to reveal their distribution patterns. Provided as an independent project through the Microbial Genomes Atlas (MiGA) web server (http://microbial-genomes.org/), the repository contains over ~2,000 genomes, more than 95% of which represent novel taxa, though representation of cultured organisms from oil-contaminated and oil reservoir ecosystems in this database is limited. The database allows researchers to classify unknown genomes by reference to known genomes, thereby facilitating the predictive understanding of microbial taxa and activities that can control the fate of oil spills.
CONCLUSIONS
Microbiology and omics tools have proven instrumental in providing an understanding of the impacts of the Deepwater Horizon oil spill on marine and coastal ecosystems. The new omics-based technologies and strategies enabled a comprehensive investigation of Gulf microbial communities and their biogeochemical functions in unprecedented detail.
Using the powerful and constantly evolving tools of genomics, GoMRI researchers discovered novel genes, pathways, organisms, communities, and partnerships associated with oil decomposition and spill remediation. Key shifts in community structures that dictate essential ecosystem functions and bioremediation services were documented. Altogether, GoMRI studies have revealed new insights and core lessons about microbial community responses to environmental disturbances, as well as their roles in maintenance and restoration of ecosystem stability during and after an oil spill. The lessons learned inform a scientific understanding of how oil spills impact various ecosystems, from deep waters to beach sands to tidal marshes, and will allow improved prediction and mitigation of damage in the event of future spills.
RESOURCES