Full Text
Ecological connectivity among coastal marine habitats—linkage in the movement of organisms and natural processes across habitat boundaries—has significant implications for the health and resilience of commercially important or threatened species in the Gulf of Maine (GOM), off the coast of the northeastern United States. Methods designed to efficiently assess connectivity are vital for identifying and managing critical habitats (Perry et al., 2018). Paired use of passive acoustic monitoring (PAM) and metabarcoding seawater samples (MSS) for observing biological and functional connectivity at various spatiotemporal scales in the marine environment is largely unexplored and may provide an efficient alternative or supplement to existing strategies.
Passive acoustic recordings are non-invasive, can identify sound sources, and can quantify spatial, temporal, and frequency attributes of an environment, collectively referred to as the soundscape. Sound sources contributing to the underwater soundscape are categorized as biological (e.g., fish vocalizations, feeding sounds), anthropogenic (e.g., vessels, underwater construction), and geophysical (e.g., breaking waves, precipitation, earthquakes). PAM enables assessment of soundscapes over extended temporal periods with minimal environmental disturbance. This permits detection of sound source presence, individual source identification, taxa detection, and quantification of properties of the acoustic environment. However, PAM is only able to detect sources that are actively producing sound and cannot confirm sound producer absence. PAM also is limited by sound recording hardware constraints, high intensity acoustic masking conditions, and generation of large quantities of data that are resource intensive to store and analyze.
Metabarcoding analyses involve extracting genetic material from particles in the environment. Using MSS, genetic material is selectively amplified with primers that target certain taxa, the amplicons are sequenced, and the reads are compared against reference databases to assign sequences to taxonomic groups. This strategy enables detection of organism presence across a broad taxonomic range. MSS has limitations, including the inability to confirm absence of taxa, biases associated with the amplification process, and diminishing DNA detectability over time.
Coupling PAM observations of biological, anthropogenic, and geophysical activity with MSS detection of taxa that may or may not produce sound provides a more complete picture of biological connectivity in terms of ecology and function. Combined characterization of soundscapes and metagenomes of similar habitat types in different geographic locations affords the opportunity to determine whether habitat type or geographic location is more influential for either platform and indicates where the strongest localized functional connectivity operates within the GOM, for example, whether sound pollution is disrupting functional connectivity among habitats.
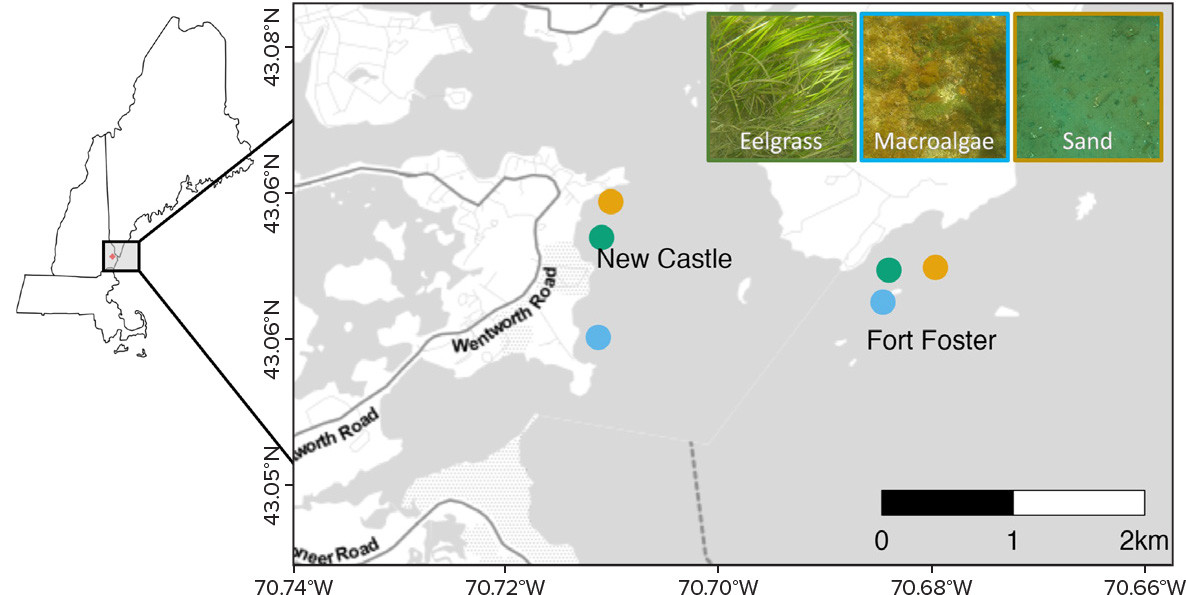
As part of a feasibility study of biological and acoustic connectivity of marine habitats, three habitat types in the shallow (<10 m depth) subtidal GOM were sampled. Target habitats, defined by biogenic and geologic components of the bottom substrate and including the water column located vertically above these substrates, were (1) eelgrass (Zostera marina) beds on soft substrates, (2) macroalgae colonizing hard substrates, and (3) soft sand/mud substrates with low biogenic cover. Two separate locations, each containing all three target habitat types, were sampled near New Castle, New Hampshire, and Fort Foster, Maine (Figure 1). These two locations were separated by ~2 km, and habitats within each were situated ≤1 km apart.
|
|
Acoustic recordings were made with SoundTrap ST500 hydrophones deployed 0.5–1 m above the substrate at all habitats. The sample rate was 144 kHz at a 50% duty cycle for a two- to three-week deployment period. Recordings collected over the deployment period were subsampled for 1 h at dawn, noon, dusk, and midnight, totaling 4 h from each day. Calculated acoustic measurements provided quantitative descriptions of the uniformity, impulsivity, and amplitude of the soundscapes in each habitat (Wilford et al., 2021). Measurements of these properties allowed for rapid, comprehensive soundscape comparisons.
During each hydrophone deployment period, 3 L water samples were collected at two- to five-day intervals near the hydrophones from 1 m above the substrate (Figure 2). These samples were stored on ice for transport to the laboratory, filtered to concentrate particles ≥5 µm, and the filters were frozen until extracted with a QIAGEN PowerWater kit. The resulting genetic material was amplified using cytochrome c oxidase subunit I (COI) and V9 primer sets that targeted metazoan and macroalgae/macrophyte DNA, respectively. Subsequently, the array of DNA sequences for each sample was determined to the genus level (Jeunen et al., 2019).
|
|
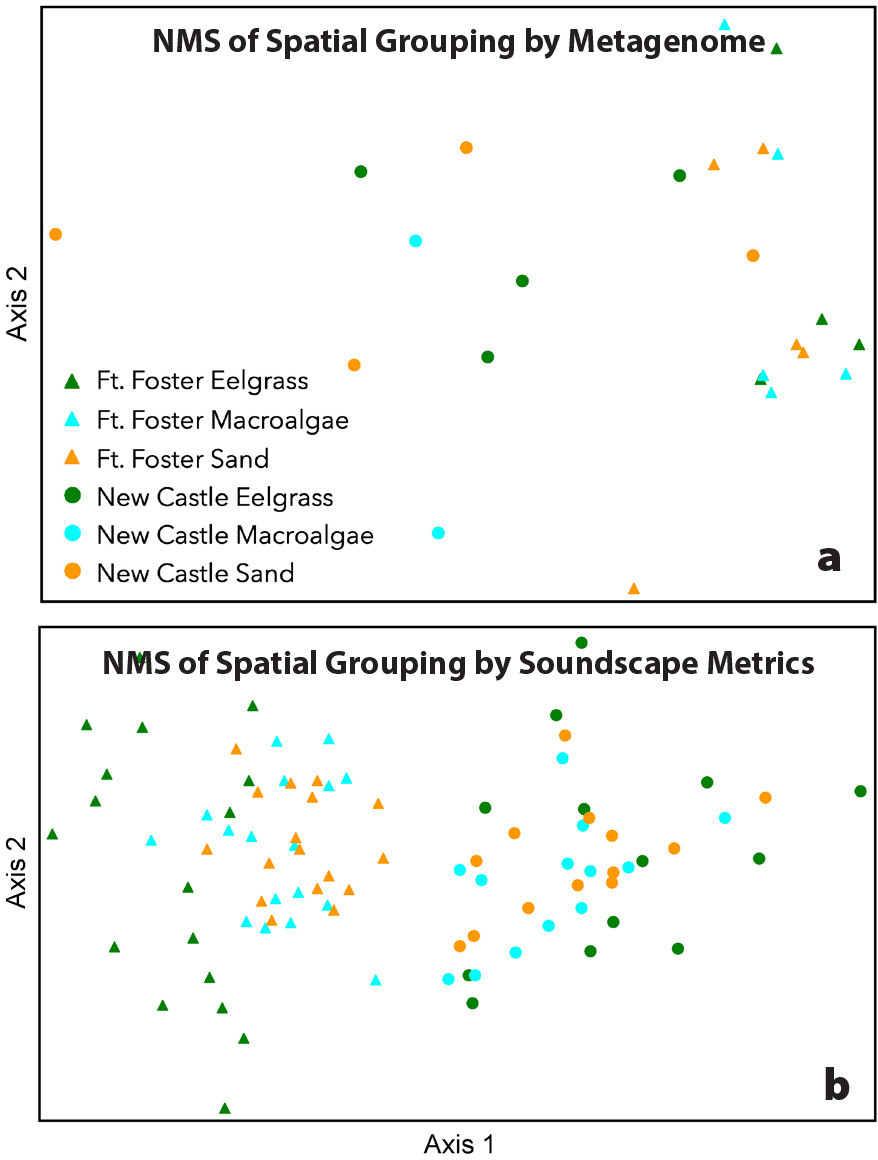
Non-metric multidimensional scaling ordination (NMS) calculated with Bray-Curtis distance for genetic and acoustic data revealed that more variation is accounted for by geographic location than by habitat type. NMS of marine taxon presence revealed clear dissimilarities among coastal metagenomes by geographic location but showed no clear grouping by habitat (Figure 3a). These preliminary results contradict previous findings that coastal habitats in New Zealand connected by water movement exhibit distinct groupings in metagenomes (Jeunen et al., 2019) potentially due to the extreme proximity and comparatively narrow taxonomic focus (metazoans, macrophytes, macroalgae) of this preliminary study, which will be expanded to include detection of additional taxa (fish, marine mammals) in future work. For soundscape measurements, NMS ordination of log10(x+1) transformed medians and central 95th percentages (C95) of each metric for each habitat showed clear grouping of soundscapes by geographic location, with some degree of separation by habitat (Figure 3b).
|
|
These preliminary results illustrate biological and functional connectivity among habitats at local scales and dissimilarity among the soundscapes and metagenomes of similar habitats at larger geographic scales. The MSS analyses complemented PAM by revealing the presence of taxa that are negligible sound producers, but that may modify the acoustic environment either by serving as sound attenuators or through association with substrates expected to differentially impact sound propagation. PAM complemented MSS by detecting anthropogenic and geophysical sound sources expected to impact the functional connectivity of habitats that would be overlooked using MSS alone. The combination of these minimally invasive observation techniques allows for more complete detection of factors contributing to both sound production and propagation than would be possible with either technique individually. Future research will involve identifying drivers of acoustic and genetic variation among geographic locations, as well as identification of indicators of similar habitat types.