Full Text
The white shark (Carcharodon carcharias) is a globally distributed, ecologically important top predator whose biology and population dynamics are challenging to study. Basic biological parameters remain virtually unknown in the Mediterranean Sea due to its historically low population density, dwindling population size, and lack of substantial sightings. White sharks are considered Critically Endangered in the Mediterranean Sea, and recent analyses suggest that the population has declined by 52% to 96% from historical levels in different Mediterranean sectors (Moro et al., 2020). Thus, white shark sightings dating back to 1860 are being used to estimate population trajectories throughout the entire region. Though the population size is unknown, remaining individuals are thought to be primarily restricted to a handful of hotspots deemed important for their reproduction and foraging. One of these hypothesized hotspots is the Sicilian Channel, which accounts for 19% of total historical sightings.
White sharks shed genetic material (skin and feces) into their environment. Non-invasive molecular methods for determining the presence of organisms in nature, such as detecting environmental DNA (eDNA) from water samples, offer a developing but powerful technique for studying cryptic or rare species. In the Mediterranean Sea, where white shark catches and sightings are limited and unpredictable, the analysis of genetic material shed from white sharks provides novel insights into their distribution and abundance, thus aiding traditional surveying methods based on encountering individuals.
Here, we describe a survey conducted in the Sicilian Channel to test and refine methods for detecting white shark eDNA and combine historical aggregations with movement simulations of eDNA molecules in seawater to interpret white shark distributions and set goals for future monitoring efforts.
White Shark eDNA Survey
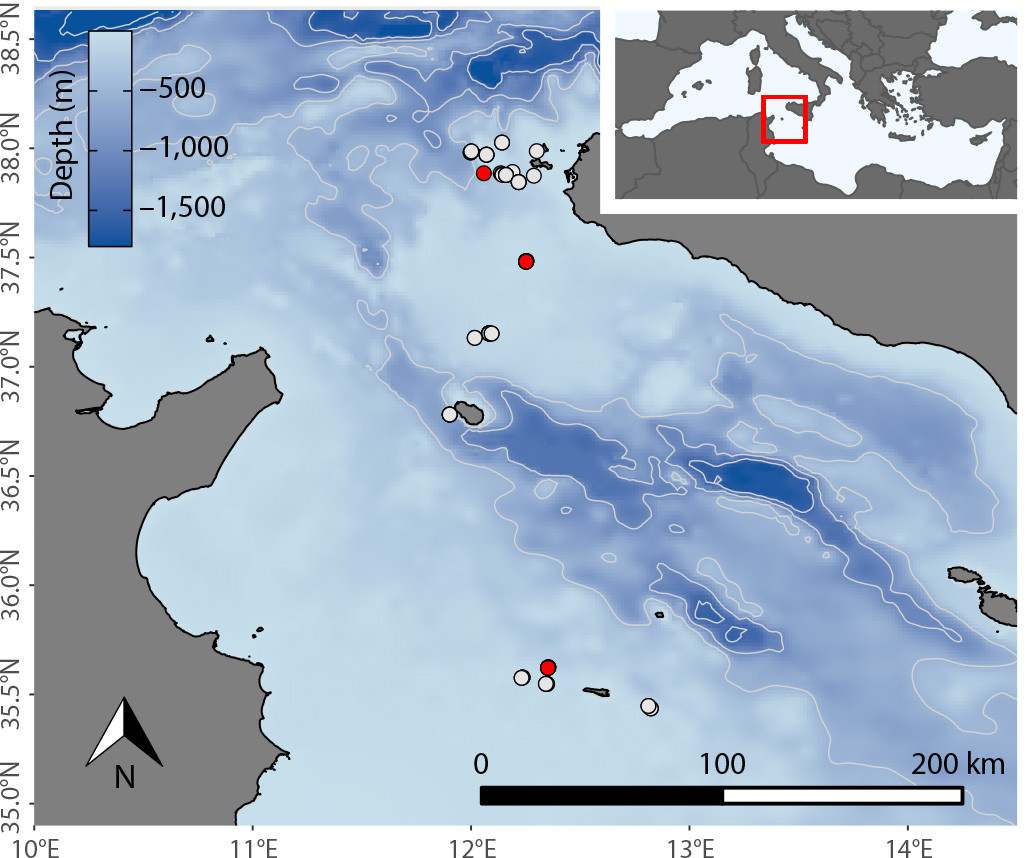
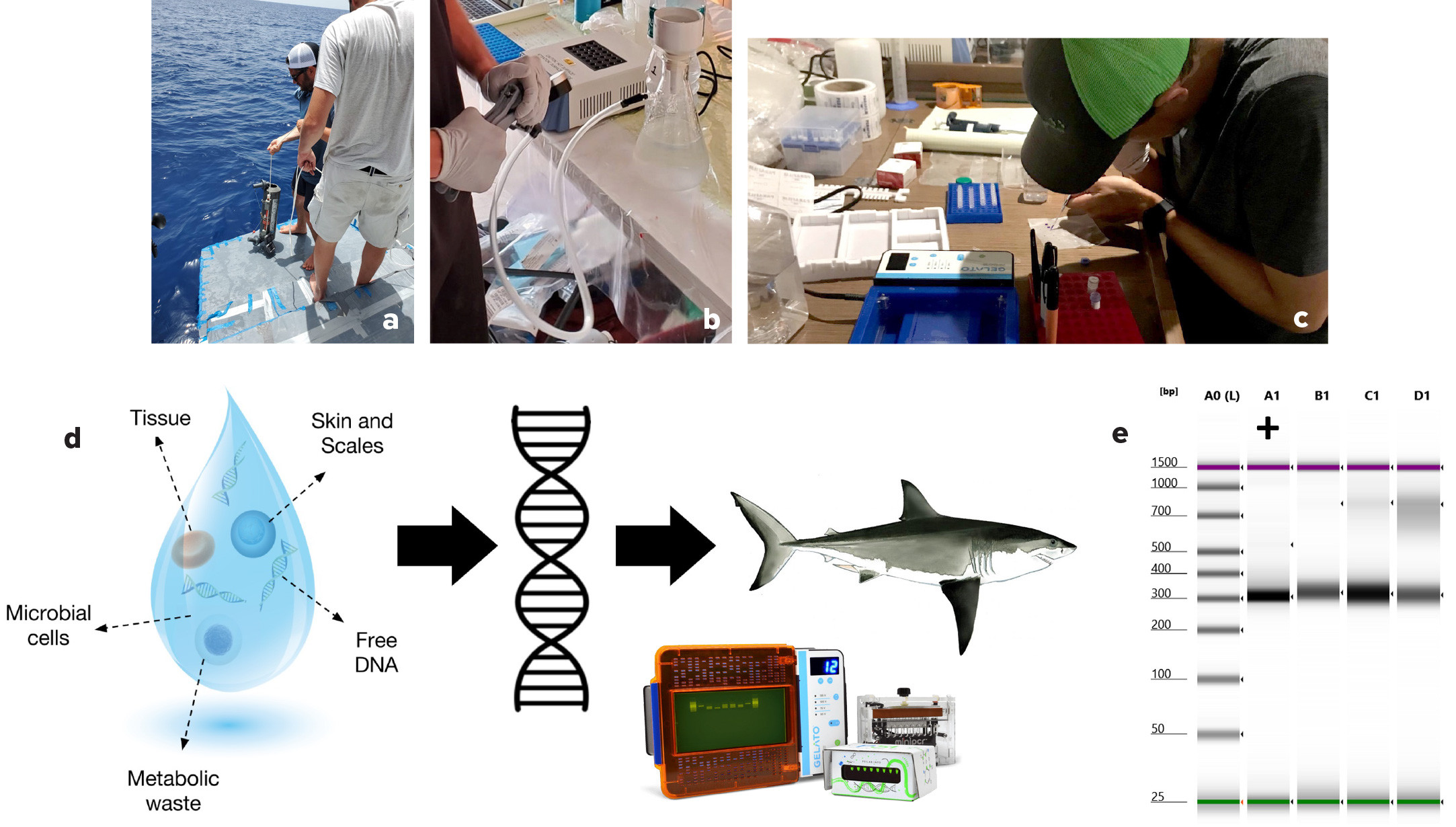
In June 2021, we collected environmental water samples from 16 sites within the Sicilian Channel (Figures 1 and 2). We filtered the samples to capture any cells and free-flowing eDNA that may be present. We amplified a unique cytochrome B gene fragment (CYTB) found in the mitochondria of white shark cells (Lafferty et al., 2018). We then observed the CYTB gene fragment on gel electrophoresis to confirm that the sample contained white shark DNA.
|
|
|
|
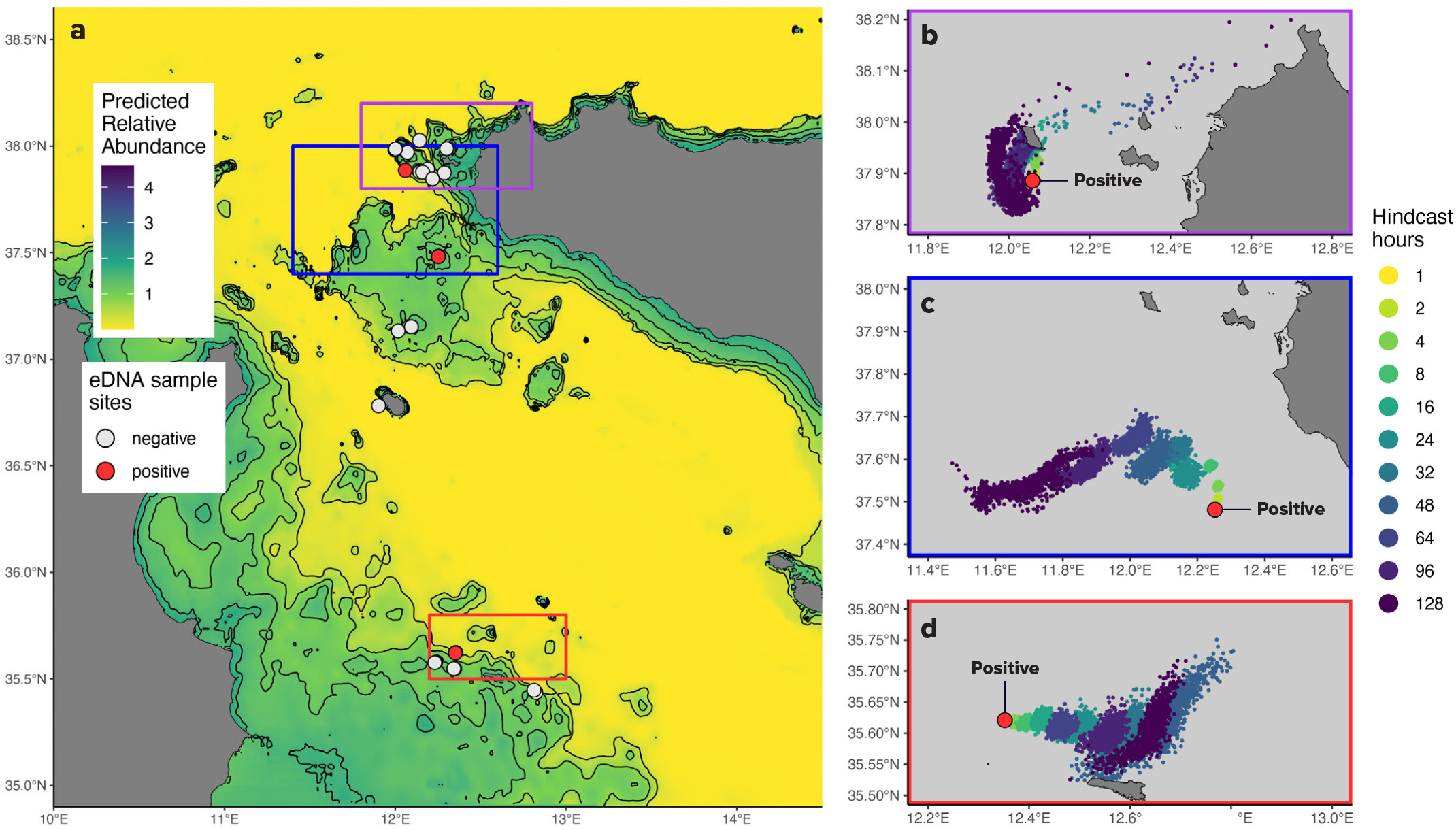
From this survey, four samples (out of 69) exhibited the unique gene fragment. These samples were selected for DNA sequencing that revealed the similarities between eDNA and the white shark reference sequence for the CYTB gene (Truelove et al., 2019). To understand the likelihood of a false-positive detection, we compared our samples to the CYTB genes of the white shark and its closest relative in the region, the shortfin mako (Isurus oxyrinchus). The four samples were significantly more similar to white shark than mako. Positive white shark detections (>95% similarity) were identified at the Pantelleria Banks and Lampedusa sites, and a third site at the Egadi Islands, indicating recent white shark presence (see online Supplementary Materials).
Using historical white shark sightings, we developed relative abundance distributions for May and June to guide our selection of survey sites (Figure 3a). By detecting the DNA of possible multiple white shark individuals, we completed an important step for confirming their presence in hypothesized hotspots. Our goal, once white shark DNA was detected, was to predict where the animal shed its genetic material by considering the movement of eDNA molecules in seawater (Figure 3b–d).
|
|
DNA molecules are subjected to environmental factors such as water currents and chemistry, tides, temperature, and sunlight once they are shed by white sharks. By modeling the trajectory of detected eDNA molecules, it is possible to predict their drifting pattern in space and time in the ocean (Andruszkiewicz et al., 2019). We ran a computer simulation to track these molecules backward in time to estimate where and when they originated. This procedure allowed us to predict the origin of eDNA from positive detections up to 128 hours prior to detection, which largely bounds the maximum time eDNA can persist in the marine realm without completely degrading to the point that it can no longer be detected (see Supplementary Materials). Hence, molecule tracking can provide insight into the locations of white shark individuals and facilitate real-time tracking of the animal to increase the chance of observing them.
These findings are congruent with our expectations to find white sharks in this specific Mediterranean sector during this season. Seasonality is known to affect not only water temperature and eDNA movement but also white shark behavior due to reproductive strategies and variation in prey abundance. Because one or multiple individuals were detected in the suspected area, these conditions were presumably favorable for white sharks. The survey produced baseline results and expectations for future monitoring of white sharks with eDNA techniques. Importantly, to construct population landscapes in the Mediterranean, there should be seasonal eDNA surveys across multiple regions.
Involving the Public in Our Work
Our approach is effective and scalable for clarifying spatiotemporal patterns in white shark distribution. To increase sampling effort and reduce ship-time costs, citizen scientists should be incorporated into eDNA surveys, as it is relatively straightforward for fishermen, tourists, and other ocean-goers to collect and filter seawater without training or prior knowledge. Expeditions are important for establishing and refining techniques, but financial and logistic constraints challenge the implementation of multiple, simultaneous sampling surveys. Therefore, public scientists are a valuable untapped resource for consistently sampling selected regions of the Mediterranean and offer the possibility for a low-cost and persistent monitoring network in parallel with scientific expeditions.
We are currently establishing a network of sailing partners in the Mediterranean Sea and have developed cost-efficient and easy-to-use kits for sampling and filtering surface water and preserving eDNA to be shipped back to our laboratory for processing. To teach citizens about the best sampling practices, we created a short instructional document and a step-by-step video to help them collect water, sterilize the equipment, and work area with diluted bleach; use latex gloves; and preserve eDNA. As the network expands, kits should be upgraded to sample at depth to help reveal the population distribution of white sharks in the Mediterranean.
In summary, detecting the DNA of Critically Endangered white shark individuals allowed us to extrapolate their spatiotemporal presence and develop a surveying strategy in a data-poor region. Expanded systematic eDNA monitoring is a promising tool for revealing the otherwise cryptic behavior of white sharks in the region. This will be crucial for developing conservation and management policies to protect the last strongholds of Mediterranean white sharks.