Full Text
At depths below 200 m, the pelagic deep sea comprises the largest, but least explored, part of the ocean. In this vast environment, animals are hard to find, and interactions among them are even harder to investigate (Robison, 2009). Climate change and industrial exploitation are exerting increasing pressure on deep-sea ecosystems, causing a decline in global ecosystem health and ecosystem services. Many of these changes occur outside of the range of human observation and are unrecognized so that effective conservation is limited. Such changes can include interactions between elusive and sometimes giant deep-sea predators such as cetaceans and their prey. The use of on-animal recorders has revealed that multiple species of cetaceans make extensive use of the deep sea, specifically the meso- (200–1,000 m depth) and bathypelagic (1,000–4,000 m depth) zones, to hunt for diverse, often cephalopod-dominated prey populations (Tyack et al., 2006). Because their dives to remote depths are energy consuming, the prey reward needs to be substantial to make the dives profitable. Thus, we expect that deep-diving cetaceans selectively target distinct foraging zones that hold specific prey communities to optimize their foraging performance.
Cephalopods are extremely abundant and play a pivotal role in marine food webs as both predators and prey (Hoving et al., 2014). For instance, it is estimated that sperm whales alone annually feed on as many cephalopods in terms of biomass as human fisheries catch worldwide. Yet deep-sea cephalopods, in particular, are understudied, and many have never been observed alive in their habitats or captured as adults (Hoving et al., 2014). Traditional methods for studying cetacean prey include net capture, optical methods, or stomach content analysis. Physical and optical sampling face the challenge that cephalopods show avoidance behavior and are patchily distributed. This results in a sampling bias toward less mobile, less sensorially equipped, and abundant specimens (Wormuth and Roper, 1983). Stomach content analysis of cetaceans is rare and requires stranding or capture, and does not typically represent a good average of the population. An alternative method for assessing regional cephalopod diversity and hence potential prey spectra of cetaceans is environmental DNA (eDNA) analysis, a relatively novel tool for studying deep-sea communities. This method exploits the phenomenon that every organism leaves genetic information in the form of DNA particles behind. These DNA particles can stem from shed cells, mucus, or feces and have the potential to reveal the identity of the source organism. eDNA analysis has been successfully used to reconstruct the horizontal distribution, diversity, and migration of open-ocean nekton (Beng and Corlett, 2020). Yet, to the best of our knowledge, eDNA analysis has not been used to investigate cephalopod biodiversity in the deep sea prior to the study presented here, which is adapted from Visser et al. (2021).
Toothed Whale Predators with Alternative Deep-Sea Feeding Grounds
We were particularly interested in the foraging behavior of Risso’s dolphins Grampus griseus and Cuvier’s beaked whales Ziphius cavirostris, two species of deep-diving toothed whales that co-occur off the Azores but exhibit two distinct deep-sea foraging strategies. Risso’s dolphins target cephalopods in epi- and mesopelagic waters, while Cuvier’s beaked whales dive deeper to meso- and bathypelagic waters to forage—indeed, the Cuvier’s beaked whale holds the record for the deepest and longest dive, which lasted more than two hours to a depth of 2,992 m (Schorr et al., 2014). Despite this segregation in hunting habitat, stomach content analyses of both species show partially overlapping prey populations dominated by deep-sea squids. A generally accepted but poorly studied hypothesis is that cetacean predators occupy different niches because they target different prey communities. This hypothesis remains largely untested, due to the methodological challenges of reaching and sampling the extreme deep-sea environment in which these animals forage.
To overcome these challenges, we pioneered the assessment of prey community composition via cephalopod eDNA analysis in combination with data on deep-sea predator foraging behavior obtained from biologging of Cuvier’s beaked whales and Risso’s dolphins. With biologging, we are tagging animals with small, animal-mounted instruments that record depth and sound to measure cetacean diving and acoustic behavior. This approach allowed us to test the hypotheses that Risso’s dolphins and Cuvier’s beaked whales exploit entirely discrete deep-sea foraging niches and that these niches hold specific prey spectra.
Cetacean Biologging
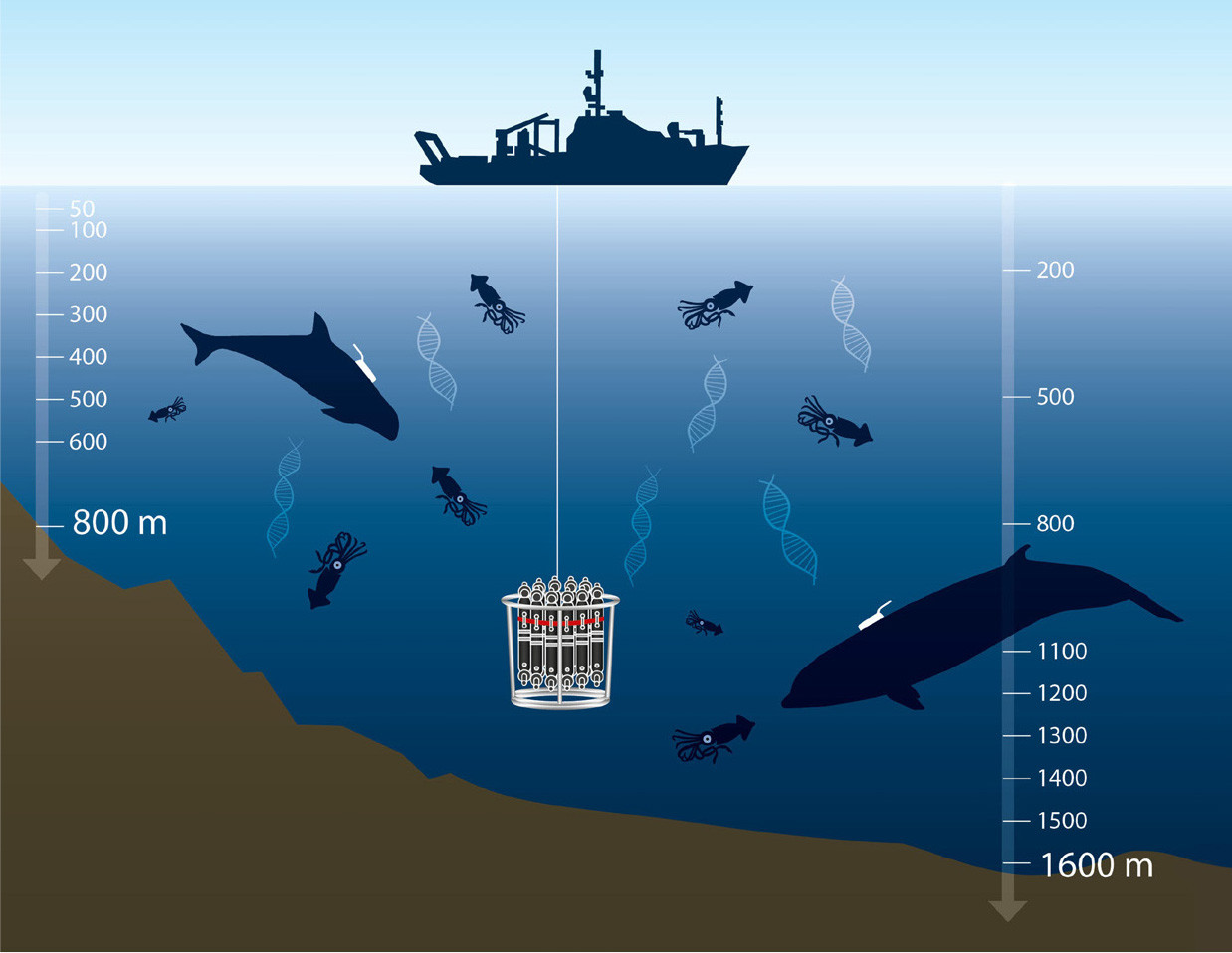
We conducted shore- and vessel-based observations to identify the foraging habitats of Cuvier’s beaked whales and Risso’s dolphins off Terceira Island, Azores, in the North Atlantic. To determine the foraging depths of the two species, individuals were tagged with noninvasive, high-resolution digital acoustic recording tags (Figure 1). The tags were attached using suction cups and released automatically. They record dive and acoustic data. Risso’s dolphins and Cuvier’s beaked whales both detect and track prey by emission of echolocation click series, so-called biosonar. Depending on the click rate and amplitude of click series emitted by the cetaceans, we can differentiate between search phases for prey (broadband click series at regular intervals) and prey capture attempts (“buzzes” when the clicks transition into discrete, rapid click series at lower amplitude). Buzzes therefore accurately indicate prey capture attempts and foraging efforts. The tag data revealed a clear niche segregation of foraging habitat and zone between Risso’s dolphins and Cuvier’s beaked whales. Risso’s dolphins hunt close to shore at depths between the surface and 600 m, while Cuvier’s beaked whales hunt further offshore at deeper depths, below 900 m in the pelagic and above the bathyal seafloor.
|
|
Cephalopod Diversity Identified from eDNA
After identifying the foraging habitats and zones of Risso’s dolphins and Cuvier’s beaked whales, we collected seawater at the respective sites throughout the water column from the surface to the seafloor (maximum depth of 1,600 m) in 100 m depth intervals (Figure 2). The collected seawater was then filtered to retrieve the eDNA. For each depth, we collected and filtered two liters in triplicate. After extracting the DNA from the filters, we amplified it via polymerase chain reaction (PCR) using two universal cephalopod primer pairs. A primer is a short piece of single-strand DNA that flanks the target region to be amplified. This target region or amplicon is later used to identify the taxa. Because a perfect primer that amplifies every species equally well does not exist, we used two primer pairs that target different gene regions to increase the number of taxa detections. We used one primer pair that targets the mitochondrial 16S rRNA gene (Jarman et al., 2006) and another that targets the nuclear 18S rRNA gene of cephalopods. The latter primer pair was developed by our lab (de Jonge et al., 2021).
|
|
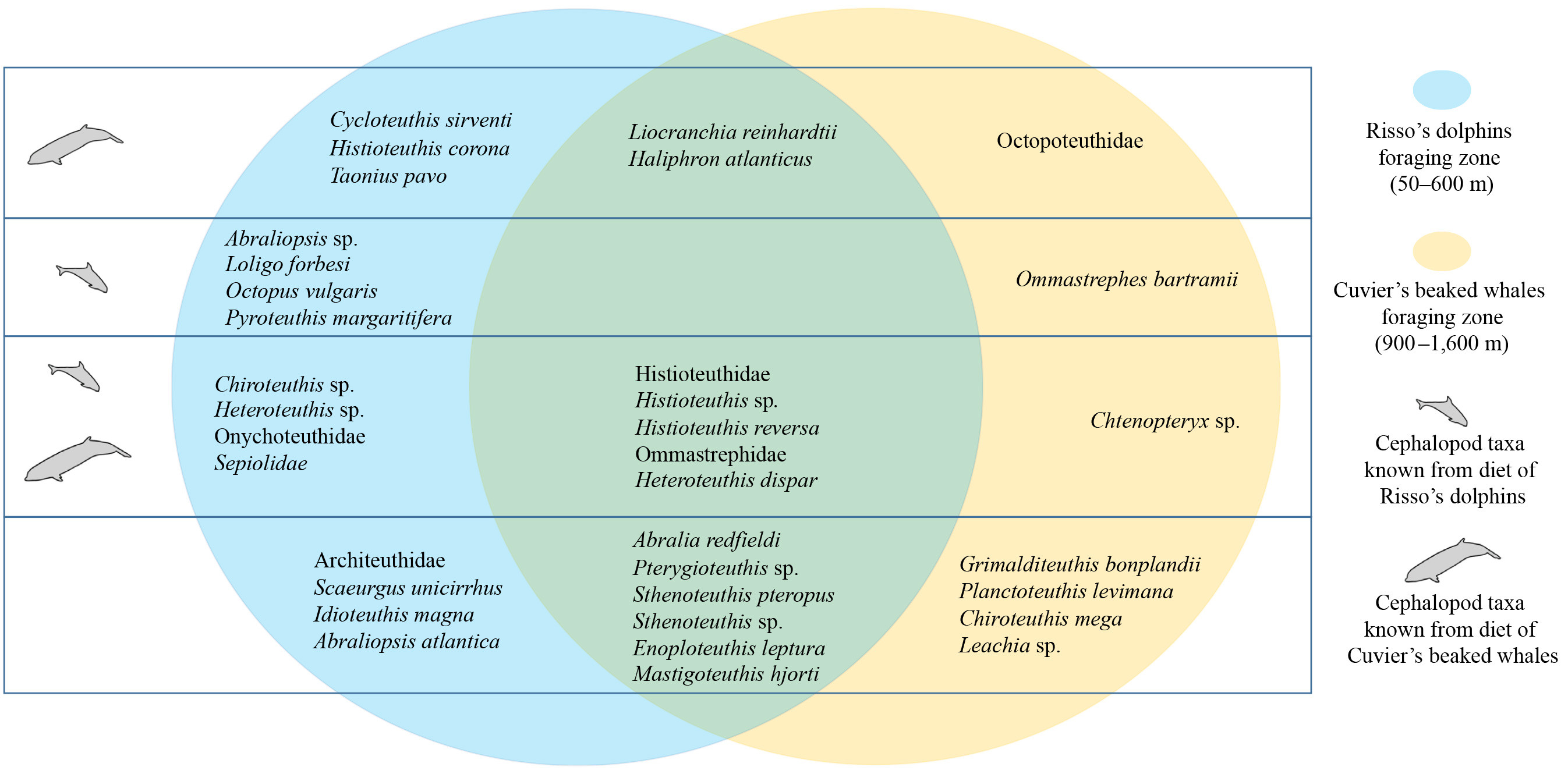
After PCR and cleaning steps, the amplified DNA was sequenced, resulting in unique genetic barcodes that can be used to identify the taxa present in the filtered water sample. Each generated eDNA barcode is then compared to a reference sequence based on a voucher specimen that has been identified. In addition to primer bias, another bottleneck in eDNA metabarcoding is the incompleteness of reference databases: if species are missing from the reference database, they cannot be identified in the eDNA data. To complement existing cephalopod reference databases, we barcoded 32 additional species that are known to occur in the North Atlantic and have been captured there in nets. Using public databases along with the one we created, we assigned taxa to the obtained eDNA sequences. As a result, we were able to identify 39 cephalopod taxa in the foraging habitats of Cuvier’s beaked whales and Risso’s dolphins, including 35 taxa that occurred in the foraging zones of both predators (Figure 3). The most widely detected taxa were Enoploteuthis leptura and Liocranchia reinhardti (Figure 4). Of the 39 cephalopod taxa, 21 could be identified to species level, which is equal to 25% of the species known to occur off the Azores. We also detected the elusive giant squid Architeuthis and two new species for this region: Chiroteuthis mega and Cycloteuthis sirventi. eDNA analysis proved to be an efficient technique for establishing diversity and distribution patterns of cephalopods in the deep sea. Our results also suggest that cephalopod eDNA is not a homogeneous mixture of DNA particles in the ocean but shows biologically meaningful distribution patterns when compared to cephalopod distributions reported in the literature. For example, strictly deep-sea taxa such as Planctoteuthis levimana and Chtenopteryx sp. were detected exclusively at great depths, while veined squid (Loligo forbesii) were only detected over the island slope at relatively shallow depths, matching its known habitat.
|
|
|
|
Deep-Sea Predator-Prey Dynamics
With biologging of Cuvier’s beaked whales and Risso’s dolphins, we were able to confirm our first hypothesis, that the two co-occurring cetaceans exploit two entirely discrete deep-sea foraging niches. By analyzing cephalopod eDNA, including use of a newly developed universal cephalopod primer pair (de Jonge et al., 2021) and the complementation of existing reference databases for cephalopods, we have provided the first reconstruction of cephalopod communities in the pelagic deep sea. Our second hypothesis, that the niches of the whales hold specific prey spectra, was not confirmed. Instead, the target zones of both cetacean species were occupied by diverse, overlapping cephalopod communities largely composed of known preferred prey. Thus, the cephalopod community composition did not explain the strict niche segregation. These findings raised the question of why Cuvier’s beaked whales dive so deep to forage when they could find the same prey species in shallower waters.
The answer came from the tagging data, which revealed that Cuvier’s beaked whales, on average, target seven to 21 fewer prey per hour than Risso’s dolphins. As Cuvier’s beaked whales are larger than Risso’s dolphins and need more energy due to their more extensive diving behavior, they must prey on larger, more calorific prey than Risso’s dolphins, which make fewer prey target attempts. eDNA analysis does not provide information on animal size, body mass, or stage of maturity. Five of the seven prey families detected through eDNA analysis, along with an additional four families in Cuvier’s beaked whale diet, are known to undergo ontogenetic migration. That is, paralarvae and juvenile cephalopods reside in surface layers to profit from increased primary productivity but descend to deeper layers as they grow to hide from predators and to reproduce (Hoving et al., 2014). Targeting different ontogenetic stages of the same prey species would reduce competition between Cuvier’s beaked whales and Risso’s dolphins. Hunting for mature and reproducing cephalopods that are more calorific than their paralarval and juvenile stages would also allow Cuvier’s beaked whales to meet their increased energetic requirements with fewer prey capture attempts. Cuvier’s beaked whales are cryptic flight strategists that form small, temporary groups with limited ability for competition or defense (Aguilar de Soto et al., 2020). Risso’s dolphins, on the other hand, are highly social and travel in large groups, which makes it easier for them to compete with other predators for resources (Hartman et al., 2008). Whereas the shallower foraging zones of Risso’s dolphins are accessible to many other cetacean species and marine top predators, resulting in increased competition, only a few predators have the ability to target prey at beaked whale foraging depths.
Interactions between cetaceans and cephalopods play a key ecological role in marine deep-sea food webs. As top predators, cetaceans may shape the population size, structure, and distribution of cephalopods. They contribute to carbon flux by predation and defecation. Combined data on potential prey spectra in foraging zones of cetaceans contribute to the understanding of marine top predator foraging behavior and of their resilience to anthropogenic noise and/or climate change. The combination of biologging and eDNA analysis demonstrated here can be applied to other predator-prey systems and help to unravel open-ocean and deep-sea food web processes.